- Title
-
Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue
- Authors
- Cao, J., Wang, J., Jackman, C.P., Cox, A.H., Trembley, M.A., Balowski, J.J., Cox, B.D., De Simone, A., Dickson, A.L., Di Talia, S., Small, E.M., Kiehart, D.P., Bursac, N., Poss, K.D.
- Source
- Full text @ Dev. Cell
|
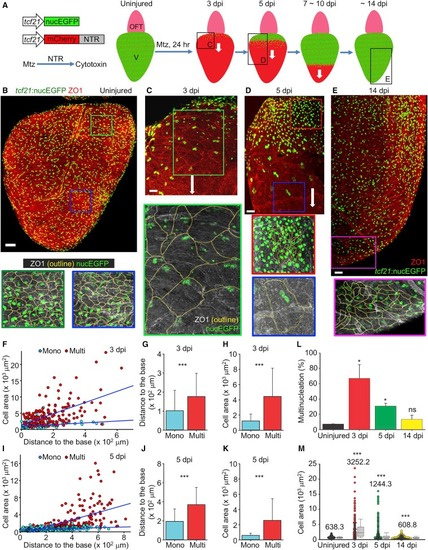
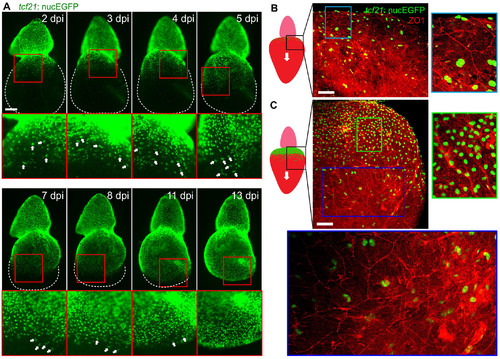
Transient Hypertrophy and Polyploidy in Regenerating Epicardial Cells
|
|
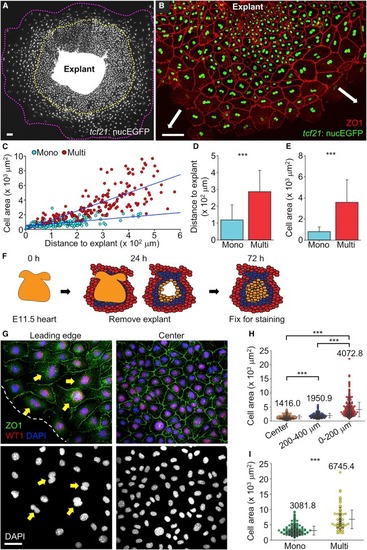
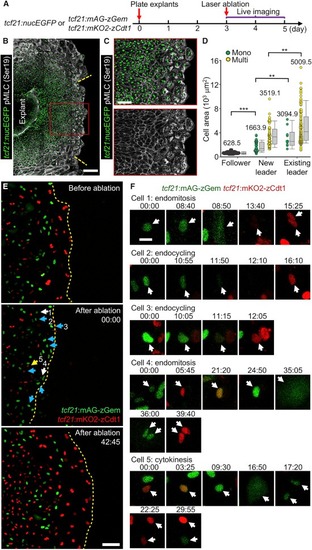
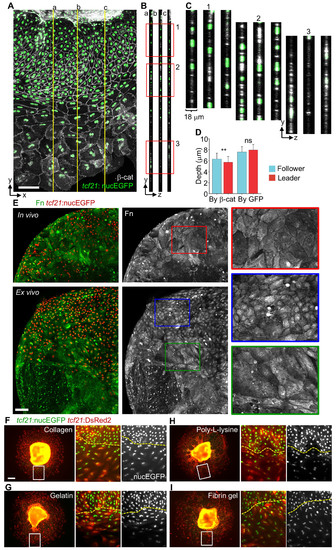
Emergence of Leader and Follower Cells Ex Vivo
|
|
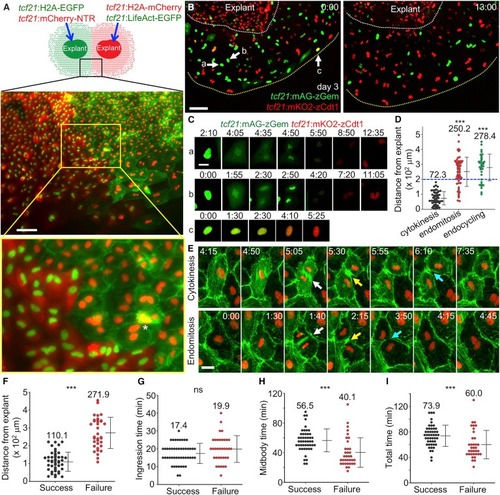
Endomitosis and Endocycling Events Underlie Epicardial Cell Polyploidy
|
|
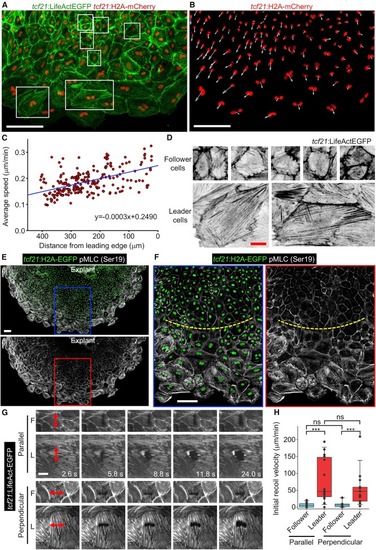
Leader Cells Display Higher Migration Velocity and Mechanical Tension than Followers
|
|
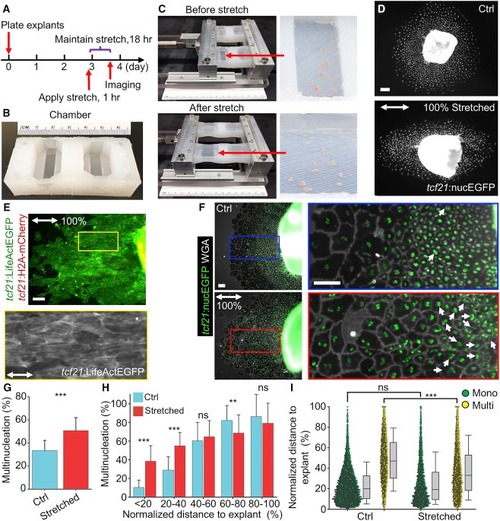
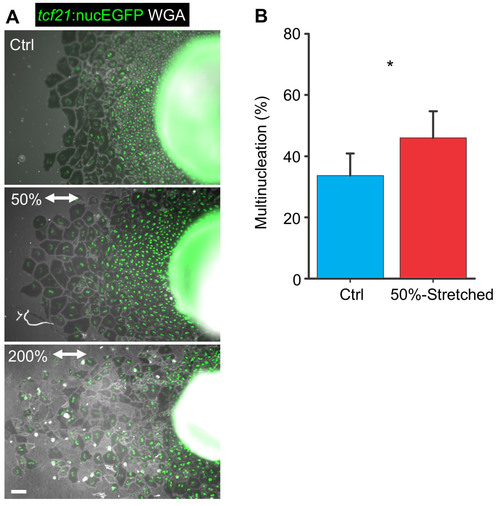
Mechanical Tissue Stretching Promotes Epicardial Endoreplication
|
|
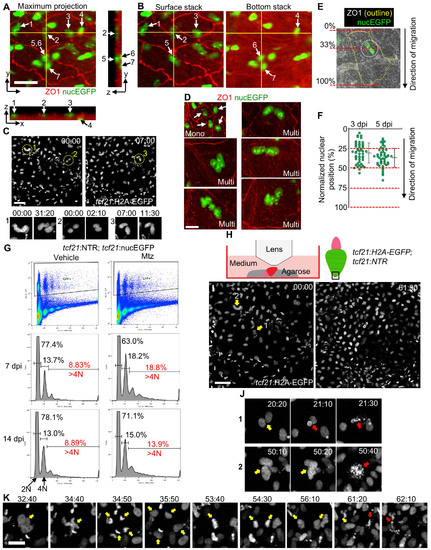
Follower Cells Undergo Endoreplication after Leader Cell Ablation
|
|
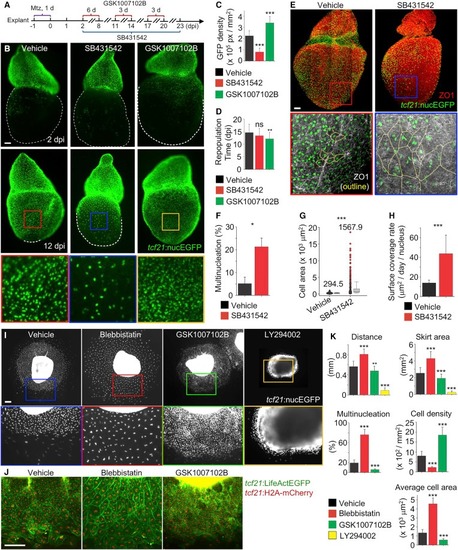
High Regenerative Capacity in Endoreplicated Cells
|
|
Analysis of Nucleation and Apoptosis in Polyploid Cells (Related to Figure 1) |
|
Cell Size and Nuclear Number during ex vivo Epicardial Regeneration (Related to Figure 1) |
|
Analysis of Cell Volume and the Relevance of ECM Components to Epicardial Regeneration (Related to Figure 2) |
|
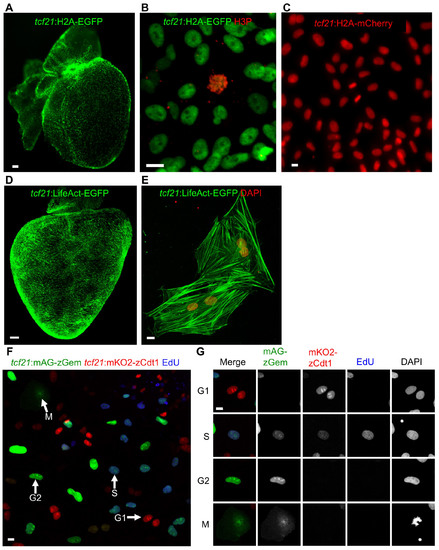
New Reporter Lines for Live Imaging (Related to Figure 3) |
|
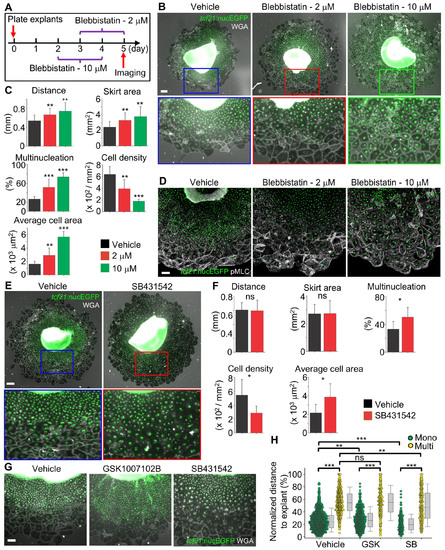
Mechanical Stretching of Epicardial Sheets (Related to Figure 5) |
|
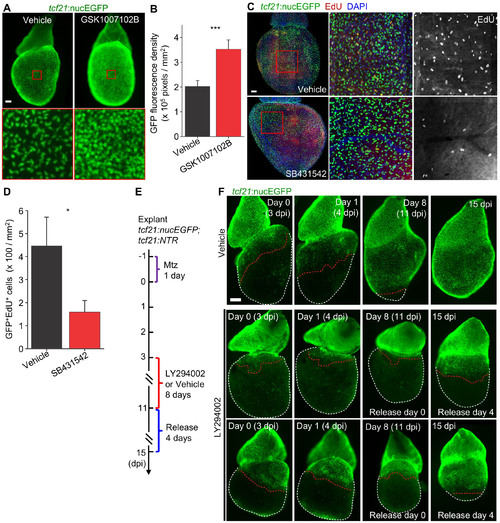
Chemical Screening for Regulators of Epicardial Cell Proliferation (Related to Figure 7) |
|
Chemical Treatment of Epicardial Explant Cultures (Related to Figure 7) |
Reprinted from Developmental Cell, 42, Cao, J., Wang, J., Jackman, C.P., Cox, A.H., Trembley, M.A., Balowski, J.J., Cox, B.D., De Simone, A., Dickson, A.L., Di Talia, S., Small, E.M., Kiehart, D.P., Bursac, N., Poss, K.D., Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue, 600-615.e4, Copyright (2017) with permission from Elsevier. Full text @ Dev. Cell