- Title
-
colgate/hdac1 repression of foxd3 expression is required to permit mitfa-dependent melanogenesis
- Authors
- Ignatius, M.S., Moose, H.E., El-Hodiri, H.M., and Henion, P.D.
- Source
- Full text @ Dev. Biol.
|
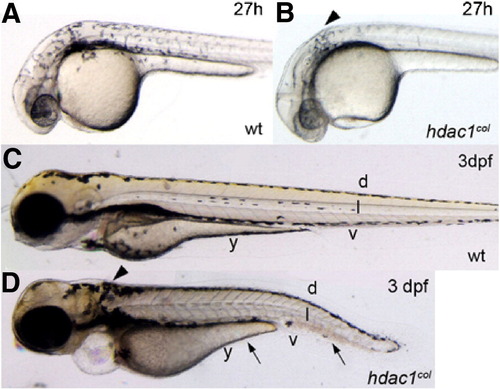
Melanophore development is defective in hdac1col mutants. Lateral views of live embryos at 27 hpf (A, B) and 3 dpf (C, D). (A, B) At 27 hpf in wild-type embryos melanophores are present posterior to the eye and otic vesicle and are migrating over the flank of the embryo. In hdac1col mutants, there are fewer melanophores and most of the melanophores are located posterior to the otic vesicle (arrowhead). There are no migrating melanophores in hdac1col mutants. (C, D) By 3 dpf, melanophores are present in four stripes in wild-type: dorsal (d), lateral (l), ventral (v) and yolk (y). In hdac1col mutants melanophore numbers do not recover. Melanophores present in hdac1col fail to migrate and are mainly localized to the dorsal stripe and a patch of melanophores posterior to the otic vesicle (arrows, arrowhead). Melanophores that do migrate ventrally in hdac1 PHENOTYPE:
|
|
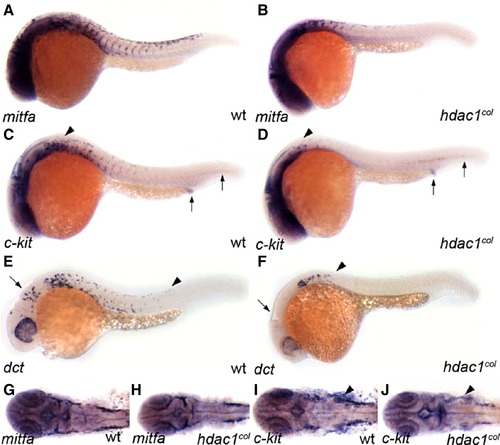
Fewer melanoblasts are specified in hdac1col embryos. Lateral and dorsal (cranial region) views of 25 hpf embryos that are stained by in situ hybridization to reveal expression of mifta (A, B, G, H), c-kit (C, D, I, J) and dct (E, F). (A, B, G, H) There are fewer mitfa-positive melanoblasts specified in hdac1col mutants as compared to wild type. (C, D) Melanoblast-specific c-kit expression in hdac1col mutants is absent and/or reduced (arrowheads), although non-melanoblast expression of c-kit in the post anal region and posterior mesoderm is equivalent to wild-type (arrows). (I, J) Similarly, in the cranial region melanoblast-specific c-kit expression is reduced or absent, while c-kit expression in the branchial arches is more robust albeit disorganized in hdac1col mutants as compared to wild-type (arrowheads). (E, F) There are fewer dct-positive differentiating melanoblasts in hdac1col mutants compared to wild-type (arrowheads) and most of the dct-positive melanoblasts are located posterior to the otic vesicle in the dorsal stripe suggesting defects in migration. Also, there is a lack of dct-positive melanoblasts in the anterior head (arrow). |
|
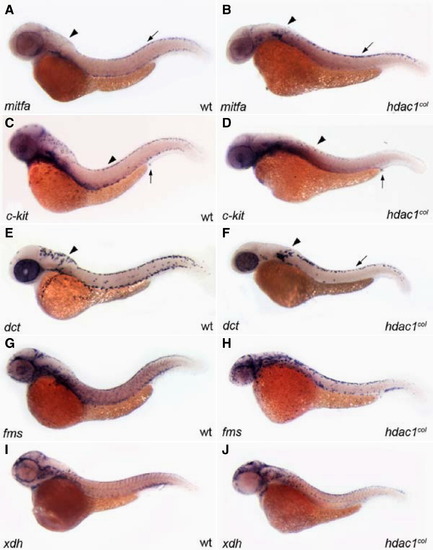
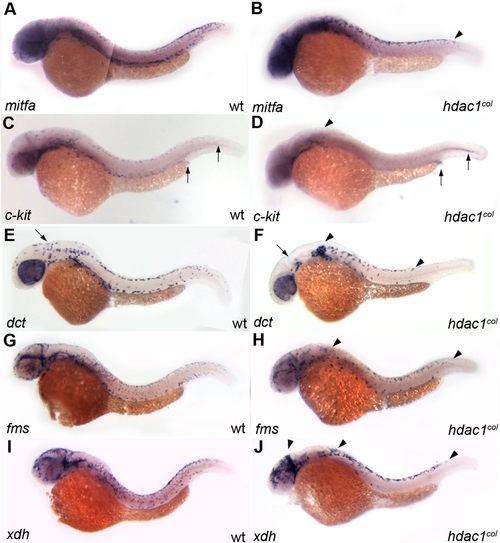
Melanophore development does not recover in hdac1col mutants. All panels are lateral views of 48 hpf embryos that are stained by in situ hybridization to reveal expression of mifta (A, B), c-kit (C, D), dct (E, F), fms (G, H) and xdh (I, J). (A, B) By 48 hpf mitfa expression is switched off in differentiating melanoblasts in wild-type; however in hdac1col mutants, mitfa continues to be expressed robustly in melanoblasts. (A, B, E, and F) There are still reduced numbers of mitfa and dct expressing melanoblasts in hdac1col mutants as compared to wild-type. Additionally, there is still a migration defect as most of the mitfa and dct expressing melanoblasts are located at their site of origin in the post otic region (arrowhead) and in the dorsal stripe (arrow). (C, D) In contrast to levels of mitfa and dct expression in melanoblasts in hdac1col mutants, c-kit expression in melanoblasts does not recover to wild-type levels (arrowheads). (G–J) Unlike melanophore development, xanthophore number and migration recover by 48 hpf, where there are numerous migrating fms-positive and xdh-positive xanthoblasts in wild-type and hdac1col mutants. |
|
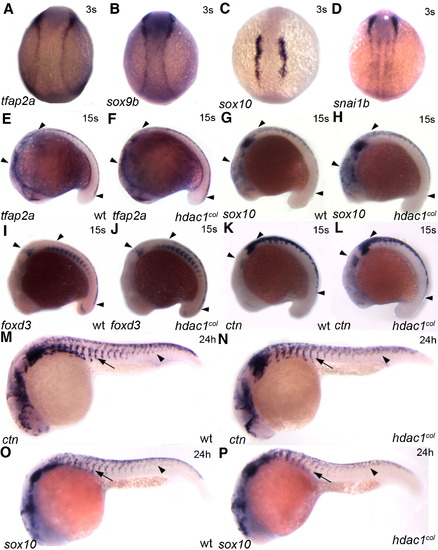
Neural crest induction and migration of non melanogenic cells are largely unaffected in hdac1col mutants. In situ hybridizations for tfap2a (A, E, F), sox9b (B), sox10 (C, G, H, O, P), snai1b (D), foxd3 (I, J) and ctn (K, L, M, N) between 3 somites (s) and 24 hpf. (A–D) Dorsal views of representative gene expression in 3 somite embryos. Neural crest induction is normal and there is no difference in the expression of tfap2a, sox9b, sox10 and snai1b between wild-type and hdac1col mutants. (E–L) Lateral views of 15 somite embryos. At the 15 s stage there is no difference in the expression and number of cranial and trunk neural crest cells (arrowheads) expressing tfap2a (E, F), sox10 (G, H), foxd3 (I, J) and ctn (K, L) between wild-type and hdac1col mutants. (M–P) Lateral views of 24 hpf embryos. Later by 24 hpf there is a slight reduction in the number of trunk neural crest cells expressing ctn and sox10 in hdac1col mutants compared to wild-type. However, the overall migration of trunk neural crest cells in hdac1col mutants is largely unaffected although slightly delayed (arrows, arrowheads). EXPRESSION / LABELING:
|
|
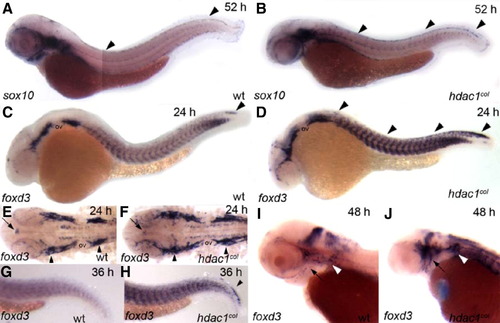
foxd3 expression is prolonged in the premigratory neural crest and increased numbers of cranial satellite glia are present in hdac1col mutants. In situ hybridizations of sox10 (A, B) and foxd3 (C–J) between 24 hpf and 52 hpf. (A, B) Lateral views of 52 hpf embryos. At 52 hpf sox10 continues to be expressed robustly in the dorsal stripe in hdac1col mutants, while expression in wild-type is absent to faint (arrowheads). (C, D) Lateral views of 24 hpf embryos. There is a prolonged expression of foxd3 in the premigratory neural crest cells at 24 hpf in hdac1col mutants compared to wild-type (D, arrowheads). In wild-type, foxd3-positive neural crest cells are mostly present at the tip of the tail (C, arrowhead). In addition to increased numbers of neural crest cells expressing foxd3, foxd3 expression is also increased in the somites in hdac1eural crest cells are mostly present at the tip of the tail (C, arrowhead). In addition to increased numbers of neural crest cells expressing %col mutants when compared to wild-type. (E, F) Flat mounts of the cranial region of 24 hpf embryos indicate that there is an increase in the number of cranial satellite glia in the pre otic and post otic placodes (arrowheads) in hdac1col mutants as compared to wild-type. In contrast, pineal gland foxd3 expression (arrows) in hdac1col mutants is slightly reduced compared to wild-type. (G, H) Lateral views of the tail region at 36 hpf. While foxd3 is not expressed in the premigratory neural crest cells in wild-type, it is still expressed in this population in hdac1col mutants (arrowhead). (I, J) Lateral views of the head at 48 hpf. The number of foxd3-positive cranial satellite glia associated with the cranial ganglia is much larger than those present in wild-type embryos (black arrow and white arrowheads). |
|
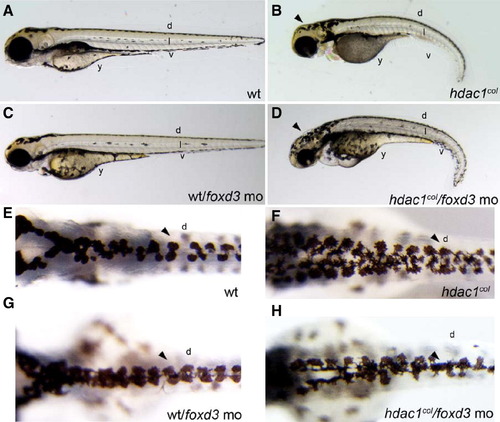
Repression of Foxd3 rescues melanogenesis in hdac1col mutants. Images of live embryos at 3 dpf, (A–D) lateral views, (E–H) dorsal views. (A, B) There are fewer melanophores present in hdac1col mutants at 3 dpf compared to wild-type embryos. Melanophores present in hdac1col mutants fail to migrate and are present at the site of origin in the post otic region, in the dorsal (d) stripe and in the anterior ventral stripe. In wild-type embryos, melanophores migrate to form four stripes; dorsal (d), lateral (l), ventral (v) and yolk (y). (C, D) Partial knockdown of Foxd3 in hdac1col mutants increases melanophore numbers and the melanophores migrate over the head (arrowhead), into the ventral stripe and over the yolk. (E, F) In control uninjected hdac1col mutants the dorsal stripe (d) is usually 3–5 melanophores wide (arrowhead), in comparison the wild-type dorsal stripe is 2ipe and over the yolk. (E, F) In control uninjected hdac1col mutants the dorsal stripe %–3 melanophores wide (arrowhead). (G, H) Partially reducing Foxd3 with a translation blocking morpholino (mo) in hdac1col mutants increases the migration of melanophores into the ventral stripe and over the yolk, the resulting dorsal stripe in hdac1col/foxd3 mo mutant/morphants is only 2–3 melanophores wide (arrowhead). PHENOTYPE:
|
|
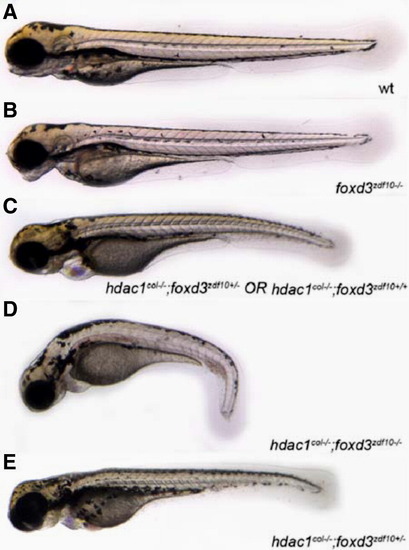
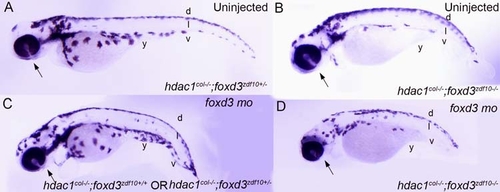
Genetically reducing foxd3 rescues melanophore development in hdac1col-/-;foxd3zdf10+/- mutants. (A–E) Live images with lateral views of 3.5 dpf embryos. Embryos obtained from a heterozygous incross of hdac1col+/-;foxd3zdf10+/- carriers gave rise to 4 distinct phenotypes. (A) Wild-type that have melanophores present in 4 distinct stripes: dorsal, lateral, ventral and yolk. (B) foxd3zdf10 mutants in which the melanophore migration patterns have largely recovered to resemble the wild-type. (C) hdac1col-/-;foxd3zdf10+/+ and hdac1col-/-;foxd3zdf10+/- mutants whose melanophore phenotype resembles single hdac1col-/- homozygous mutant phenotype. (D) hdac1col-/-;foxd3zdf10-/- double mutants in which there is a reduction in melanophore numbers as well as reduced migration into the lateral, ventral and yolk stripe locations. (E) 10% of phenotypically hdac1col mutants that are hdac1col-/-;foxd3zdf10+/- display a rescue of melanophore number as well as migration into the lateral, ventral and yolk stripes. |
|
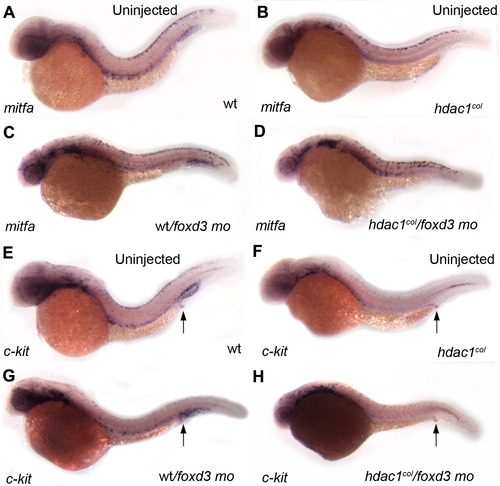
Foxd3 negatively regulates mitfa. (A–D) Lateral views of mitfa in situ hybridizations of 32 hpf embryos. (B, D) There is an increase in the number of mitfa-positive melanoblasts in hdac1col/foxd3 mutant/morphant embryos as compared to uninjected hdac1col mutants. (A, C) In wild-type/foxd3 morphant embryos mitfa expression in melanoblasts is more robust than uninjected controls. (E–H) Lateral views of c-kit in situ hybridizations in 32 hpf embryos. (F, H) There is no rescue of c-kit expression in melanoblasts in hdac1col/foxd3 mutant morphants when compared to uninjected hdac1col mutants at 32 hpf. (E, G) Qualitatively, there are fewer c-kit positive melanoblasts in foxd3 morpholino treated wild-type embryos as compared to uninjected wild-type embryos. (E–H) In contrast to melanoblast expression, there is no difference in c-kit expression in the post anal region between wild-type, hdac1col mutants, wt/foxd3 wild-type/morphants and hdac1col/foxd3 mutant/morphant embryos (arrows). EXPRESSION / LABELING:
|
|
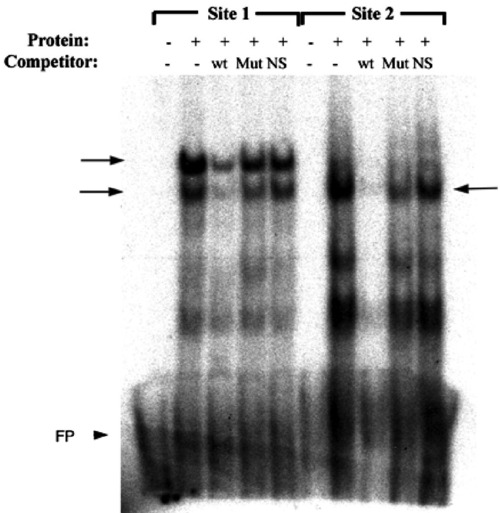
Foxd3 can physically interact with predicted forkhead binding sites in the mitfa promoter. Electrophoretic mobility shift assay (EMSA) using synthetic Foxd3 protein, radiolabeled conserved region probes spanning the predicted forkhead binding sites (Site 1 and Site 2), and unlabeled wild-type or mutated site 1 or site 2 or unrelated (OCTA) oligonucleotide competitors. Arrows denote specific bands observed with Foxd3 protein. wt—wild-type site 1 or site 2 competitors; Mut—mutated site 1 or site 2 competitors; NS—nonspecific (OCTA) competitor; FP—free probe (arrowhead). |
|
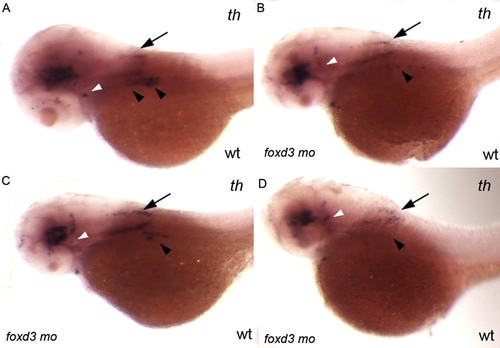
Injection of 0.56 ng of foxd3 morpholino per embryo only partially phenocopies the foxd3zdf10 mutant sympathetic neuron defect. A–D: Side views of 56 hpf wild-type embryos labeled by in situ hybridization expression for tyrosine hydroxylase. foxd3 is required for sympathetic neuron development and loss of foxd3 in foxd3zdf10 mutants or by knockdown with high doses (20 ng/embryo) of translational blocking morpholino results in the complete loss of th-positive sympathetic neurons. A: Wild-type uninjected control embryo in which there are two clear stripes of th-positive sympathetic neurons in the anterior trunk region (arrowheads). B–D: All embryos shown are from the same treated clutch. Reducing Foxd3 partially (0.56 ng/embryo) decreases the number of th-positive sympathetic neurons, although rare wild-type morpholino treated embryos (D) have an absence of sympathetics (arrowheads). A–D: In contrast to differences in th-positive sympathetic neuron expression, th expression in the hind-brain (arrows) and locus coeruleus (white arrowheads) is similar between control uninjected and wild-type/foxd3 morphants. |
|
Melanoblast differentiation and migration do not recover at 30 hpf. All panels are lateral views of 30 hpf embryos that are stained by in situ hybridization to reveal expression of mifta (A, B), c-kit (C, D), dct (E, F), fms (G, H) and xdh (I, J). A, B: By 30 hpf, the number of melanoblasts that express mitfa does not recover in hdac1col mutants. Also, most of the mitfa-positive melanoblasts fail to migrate and are located in the dorsal stripe and in the post otic region in hdac1col mutants. In contrast, wild-type melanoblasts have migrated into the ventral stripe and over the head. C, D: Additionally, melanoblast-specific c-kit expression in hdac1col is still absent and/or reduced (arrowhead), although non-melanoblast expression of c-kit in the post anal region and posterior mesoderm is equivalent to wild-type (arrows). E, F: There are fewer dct-positive differentiating melanoblasts in hdac1col mutants as compared to wild-type (arrowheads) and most of the dct-positive melanoblasts are located posterior to the otic vesicle and in the dorsal stripe. A few dct-positive melanoblasts are migrating in the anterior trunk in hdac1col mutants, although dct is not expressed as robustly compared to expression in wild-type melanoblasts. G, H: In contrast to c-kit-positive melanoblasts, there are many more fms-positive differentiating xanthoblasts in hdac1col mutants compared to wild-type. I, J: Although reduced in number when compared to wild-type, xdh-positive differentiating xanthoblasts are more numerous in the cranial region (arrows) and trunk (arrowheads) as compared to differentiating dct-positive melanoblasts in hdac1col mutants. EXPRESSION / LABELING:
|
|
Further repression of Foxd3 in hdac1col-/-;foxd3zdf10+/- and hdac1col-/-;foxd3zdf10+/+ mutants increases the rate of rescue of melanogenesis. A-D: Side views of live images of hdac1col embryos at 3.5 dpf obtained from hdac1col+/-;foxd3zdf10+/- heterozygous mutant incrosses in which either Foxd3 is further reduced with 0.477 ng of foxd3 morpholino (mo) or are uninjected controls. A: At 3.5 dpf 10% of hdac1col embryos obtained from a double mutant hdac1col+/-;foxd3zdf10+/- heterozygous incross have increased number of melanophores which migrate over the head, into the ventral stripe (v) in the tail and onto the yolk sac areas (y). The genotype of the rescued mutants was identified as hdac1col-/-;foxd3zdf10+/-. B: hdac1col-/-;foxd3zdf10-/- double homozygous mutants have fewer melanophores than hdac1col-/- single mutants and most of the melanophores are present in the dorsal (d) stripe. C, D: Reducing Foxd3 via morpholino rescues the melanophore defect in hdac1col-/-;foxd3zdf10+/- and hdac1col-/-;foxd3zdf10+/+ mutants (62.5% of all hdac1col mutants) but not in hdac1col-/-;foxd3zdf10-/- double homozygous mutants. A-D: hdac1col-/-;foxd3zdf10+/-, hdac1col-/-;foxd3zdf10+/+ and hdac1col-/-;foxd3zdf10-/- mutants have the same eye defect as hdac1col-/- single mutants and knockdown of Foxd3 has no effect on the eye phenotype (arrows). PHENOTYPE:
|

Unillustrated author statements |
Reprinted from Developmental Biology, 313(2), Ignatius, M.S., Moose, H.E., El-Hodiri, H.M., and Henion, P.D., colgate/hdac1 repression of foxd3 expression is required to permit mitfa-dependent melanogenesis, 568-583, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.