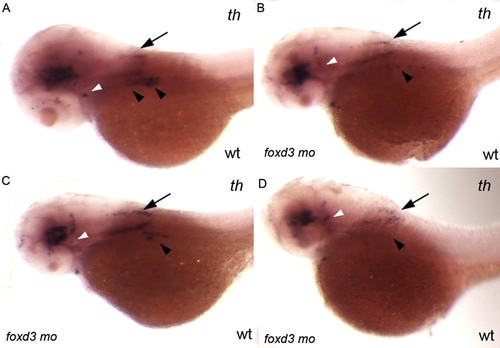
Injection of 0.56 ng of foxd3 morpholino per embryo only partially phenocopies the foxd3zdf10 mutant sympathetic neuron defect. A–D: Side views of 56 hpf wild-type embryos labeled by in situ hybridization expression for tyrosine hydroxylase. foxd3 is required for sympathetic neuron development and loss of foxd3 in foxd3zdf10 mutants or by knockdown with high doses (20 ng/embryo) of translational blocking morpholino results in the complete loss of th-positive sympathetic neurons. A: Wild-type uninjected control embryo in which there are two clear stripes of th-positive sympathetic neurons in the anterior trunk region (arrowheads). B–D: All embryos shown are from the same treated clutch. Reducing Foxd3 partially (0.56 ng/embryo) decreases the number of th-positive sympathetic neurons, although rare wild-type morpholino treated embryos (D) have an absence of sympathetics (arrowheads). A–D: In contrast to differences in th-positive sympathetic neuron expression, th expression in the hind-brain (arrows) and locus coeruleus (white arrowheads) is similar between control uninjected and wild-type/foxd3 morphants.
|

