- Title
-
Single-cell profiling and zebrafish avatars reveal LGALS1 as immunomodulating target in glioblastoma
- Authors
- Finotto, L., Cole, B., Giese, W., Baumann, E., Claeys, A., Vanmechelen, M., Decraene, B., Derweduwe, M., Dubroja Lakic, N., Shankar, G., Nagathihalli Kantharaju, M., Albrecht, J.P., Geudens, I., Stanchi, F., Ligon, K.L., Boeckx, B., Lambrechts, D., Harrington, K., Van Den Bosch, L., De Vleeschouwer, S., De Smet, F., Gerhardt, H.
- Source
- Full text @ EMBO Mol. Med.
|
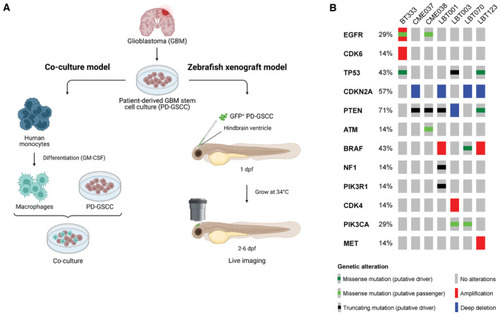
Overview experimental set‐up and included GSCCs Schematic overview of the study design. Eight different GSCCs were used in a co‐culture model with human monocyte‐derived macrophages (left) and in an orthotopic zebrafish xenograft model (right). dpf, days post fertilization. Oncoprint of included GSCCs: broad spectrum of common genetic aberrations in GBM were covered. Included GSCCs are mentioned on top of the figure. |
|
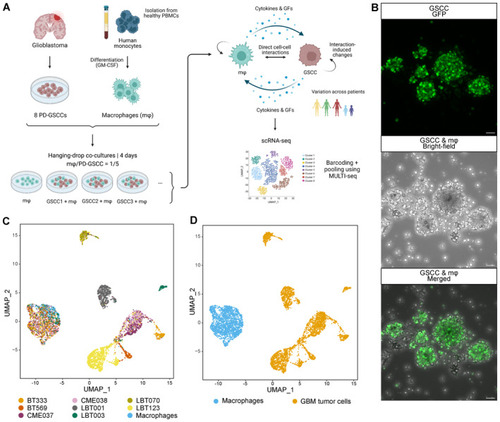
Single‐cell profiling of GSCC‐macrophage co‐cultures Schematic overview of the experimental set‐up of the scRNA‐seq profiling assay. PBMCs, peripheral blood mononuclear cells; GFs, growth factors. High‐resolution images of a GSCC‐macrophage co‐culture. GFP+ GSCC was used for visualization of the co‐culture with non‐labeled macrophages. Scale bars: 50 μm. Uniform Manifold Approximation and Projection (UMAP) plots of 5,320 cells from nine samples, annotated by sample name (C) and by cell type (D). |
|
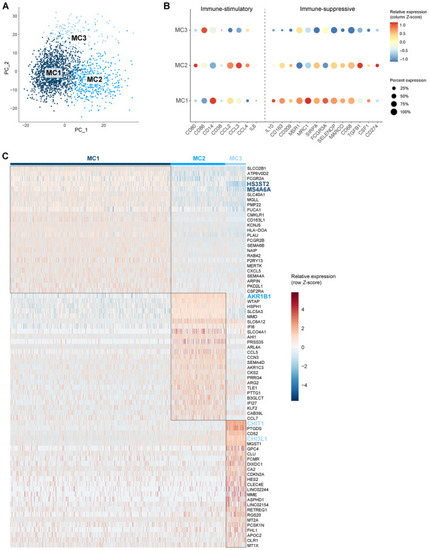
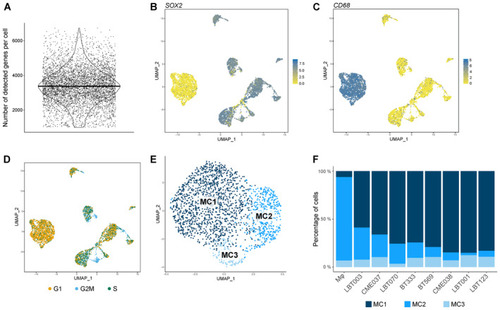
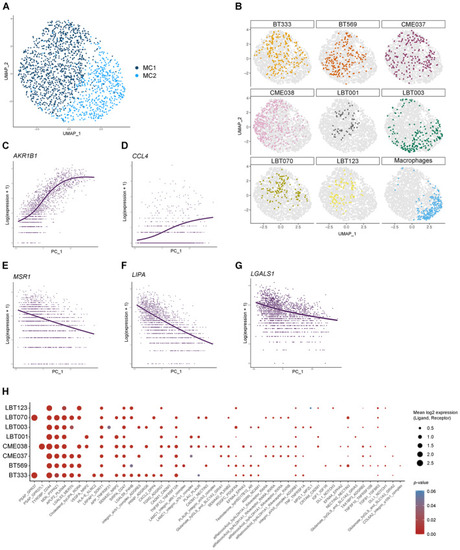
Molecular heterogeneity of GBM‐associated macrophages Principal component analysis (PCA) plot of macrophage population identified three distinct macrophage subclusters (MC1‐3). PCs were calculated using the 2,000 most variable genes. Plot shows PC1 and PC2. Dot plot showing marker gene expression for immune‐stimulation and ‐suppression. Dot size indicates the percentage of cells in each macrophage subcluster expressing the gene, and dot color indicates the relative expression level. Heatmap of top 25 differentially expressed genes in the macrophage subclusters, ranked by log2(FC). Genes discussed in the text are highlighted in the subcluster colors. |
|
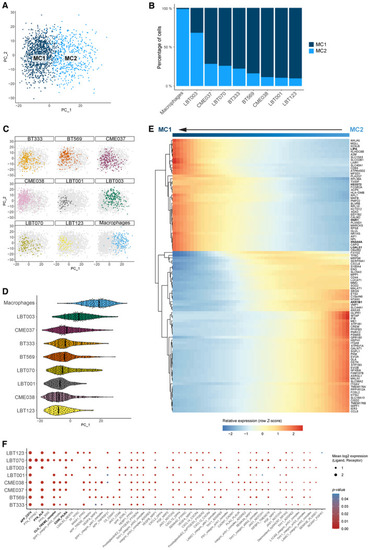
Macrophages shift toward an immunosuppressive phenotype upon co‐culture with patient‐derived GSCCs PCA plot of macrophage population without TransMos shows two distinct macrophage subclusters (MC1‐2). PCs were calculated using the 2,000 most variable genes. PC1 and PC2 are shown on the PCA plot. Macrophage subcluster distribution for the different samples. Representation of original samples on the PCA plot. Pseudotime analysis along PC1 axis. Violin plots depicting the PC1 values of each single cell, split up by sample. Heatmap of the 100 most significant temporally expressed genes along the PC1 axis, constructed by fitting a generalized additive model with the PC1 value as a LOESS term (see Dot plot of cell–cell communication analysis using CellPhoneDB. Depicted are L:R pairs for GSCC ‐ macrophage signaling across all GSCCs, ranked by mean log2 expression. Each dot size shows the log2 mean of expression values and dot color indicates the |
|
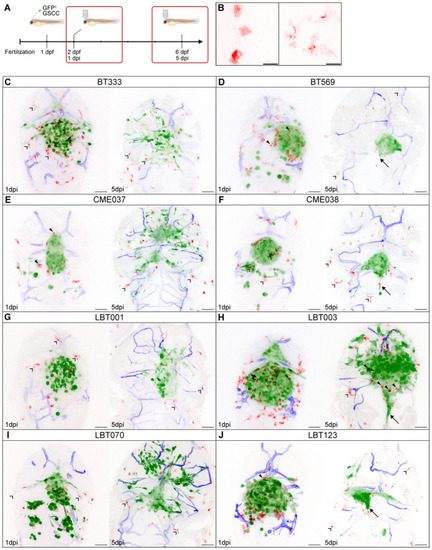
Overview of different zebrafish avatars Schematic overview showing the timeline of the orthotopic zebrafish xenograft model. Zoomed‐in images of round (left) and ramified (right) GAMs in the recorded time‐lapse movies. Scale bars: 15 μm. Representative maximum intensity projections of a z stack of the head region of |
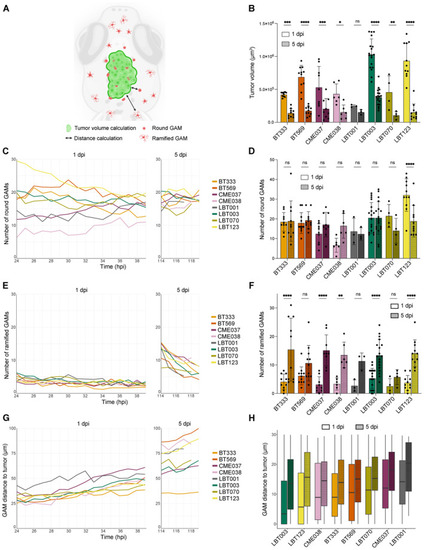
|
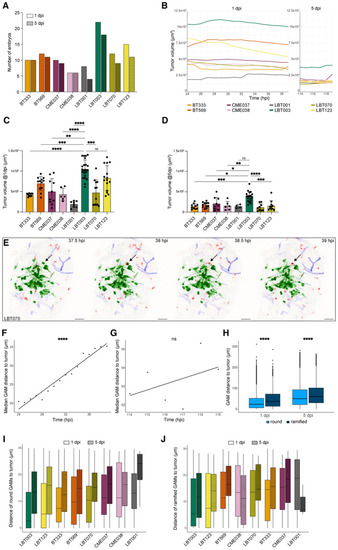
GSCC‐specific morphometrics and dynamics of the tumor and its microenvironment in 3D over time Schematic overview of features extracted and computed from the time‐lapse movies of the orthotopic zebrafish xenograft model. Tumor volume at the start of 1 and 5 dpi time‐lapse movies ( Mean number of round GAMs over time, during 1 dpi movies (left) and 5 dpi movies (right) ( Number of round GAMs at the start of 1 and 5 dpi time‐lapse movies ( Mean number of ramified GAMs over time, during 1 dpi movies (left) and 5 dpi movies (right) ( Number of ramified GAMs at the start of 1 and 5 dpi time‐lapse movies ( Mean GAM distance to the tumor over time, during 1 dpi movies (left) and 5 dpi movies (right) ( Boxplot of GAM distance to the tumor of all GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies, ranked by increasing median distance at 1 dpi ( Data information: Data describe biological replicates, i.e. individual zebrafish embryos. The xenograft experiment was replicated once for CME038, twice for BT333, BT569, CME037, LBT001, three times for LBT070 and LBT123, and four times for LBT003. In (B, D and F), data are presented as mean ± SD. The |
|
Macrophage/GAM‐GSCC interactions correlate to clinical outcome in GBM patients Swimmer plot of included patients with indication of important events. UMAP plot of GBM tumor cells annotated by sample name. Two‐dimensional butterfly plot visualization of molecular subtype signature scores according to Neftel Dot plot showing the 25 most significant positive and negative enriched GSEA pathways using all curated gene sets of WikiPathways, Reactome, KEGG, PID and BioCarta databases for the group of invasive GSCCs (cut‐off corrected Correlogram of |
|
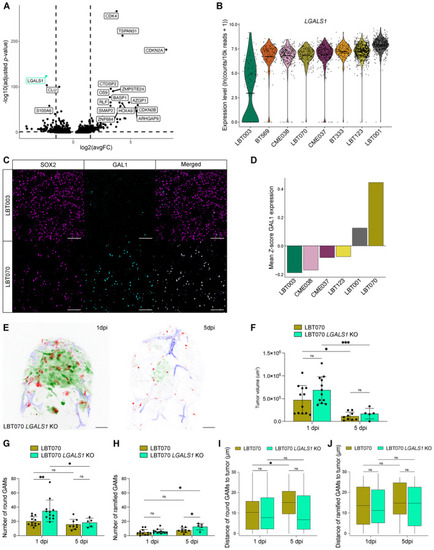
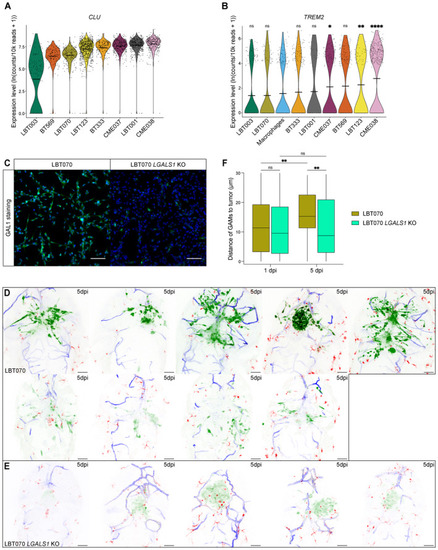
Volcano plot depicting differentially expressed genes in LBT003 and all other GSCCs (Left: downregulated genes in LBT003; right: upregulated genes in LBT003). Violin plot showing Representative double immunofluorescence images showing co‐expression of SOX2 (magenta) and GAL1 (cyan) in GBM tissue from LBT003 and LBT070. For enhanced visualization, a binary mask was generated from the SOX2+ cells and multiplied with the image of GAL1 staining in Fiji to exclude GAL1 staining in non‐tumor cells. Scale bars: 100 μm. Mean fluorescence intensity values for GAL1 staining in SOX2+ cells in GBM tissue samples, normalized using Representative maximum intensity projections of a z stack of the head region of a Tumor volume at the start of 1 and 5 dpi time‐lapse movies ( Number of round GAMs at the start of 1 and 5 dpi time‐lapse movies ( Number of ramified GAMs at the start of 1 and 5 dpi time‐lapse movies ( Boxplot of GAM distance to the tumor of round GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies ( Boxplot of GAM distance to the tumor of ramified GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies ( Data information: The zebrafish experiments describe biological replicates, i.e. individual zebrafish embryos. The xenograft experiment was replicated twice for LBT070 |
|
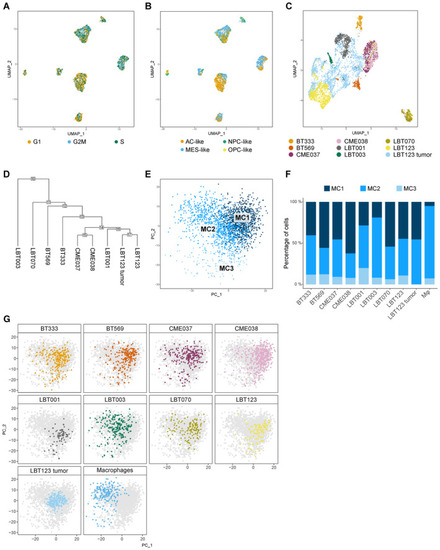
Single‐cell profiling of GSCC‐macrophage co‐cultures reveals molecular heterogeneity of GBM‐associated macrophages Violin plot showing the number of detected genes per cell ( UMAP plots showing UMAP plot showing cell cycle score. Proliferating GBM tumor cells are depicted in blue. UMAP plot of macrophage population identified three distinct macrophage subclusters (MC1‐3). Macrophage subcluster distribution for the different samples. m |
|
Macrophages shift toward an immunosuppressive phenotype upon co‐culture with patient‐derived GSCCs UMAP plot of macrophage population without TransMos shows two distinct macrophage subclusters (MC1‐2). Representation of original samples on the UMAP plot. LOESS plots for Dot plot of cell–cell communication analysis using CellPhoneDB. Depicted are L:R pairs for macrophage ‐ GSCC signaling across all GSCCs, ranked by mean log2 expression. Each dot size shows the log2 mean of expression values and dot color indicates the |
|
GSCC‐specific morphometrics and dynamics of the tumor and its microenvironment in 3D over time Number of zebrafish embryos used for the time‐lapse movies that were generated at 1 and 5 dpi. Mean tumor volume over time, during 1 dpi movies (left) and 5 dpi movies (right) ( Tumor volume at the start of 1 dpi time‐lapse movies ( Tumor volume at the start of 5 dpi time‐lapse movies ( Representative maximum intensity projections of a z stack of the head region of a Trend line of median GAM distance to the tumor over time in 1 dpi (F) and 5 dpi (G) movies for all GSCCs ( Boxplot of distance of round and ramified GAMs to the tumor for all GSCCs, at the start of 1 and 5 dpi time‐lapse movies ( Boxplot of GAM distance to the tumor of round GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies, ranked by increasing median distance at 1 dpi ( Boxplot of GAM distance to the tumor of ramified GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies, ranked by increasing median distance at 1 dpi ( Data information: Data describe biological replicates, i.e. individual zebrafish embryos. The xenograft experiment was replicated once for CME038, twice for BT333, BT569, CME037, LBT001, three times for LBT070 and LBT123, and four times for LBT003. In (C and D), data are presented as mean ± SD. In (C), the |
|
Macrophage/GAM‐GSCC interactions correlate to clinical outcome in GBM patients UMAP plot of GBM tumor cells showing cell cycle score. UMAP plot of GBM tumor cells showing Neftel subtypes. UMAP plot of GBM tumor cells, including cells from the original LBT123 tumor. Clustering tree of GBM tumor cells. The numbers in the boxes at the nodes indicate the order in which the clusters were merged during the hierarchical clustering process. The internal nodes, which start at the number of leaf nodes + 1, represent the nodes that separate groups at different levels of the hierarchical clustering. PCA plot of Macrophage/GAM subcluster distribution for the different samples, including cells from the original LBT123 tumor. m Representation of original samples on PCA plot, including the original LBT123 tumor. |
|
Violin plot showing Violin plot showing Representative immunofluorescence images of LBT070 Maximum intensity projections of a z stack of the head region of Boxplot of GAM distance to the tumor of all GAMs within 30 μm of the tumor, at the start of 1 and 5 dpi time‐lapse movies ( Data information: The zebrafish experiments describe biological replicates, i.e. individual zebrafish embryos. The xenograft experiment was replicated twice for LBT070 |