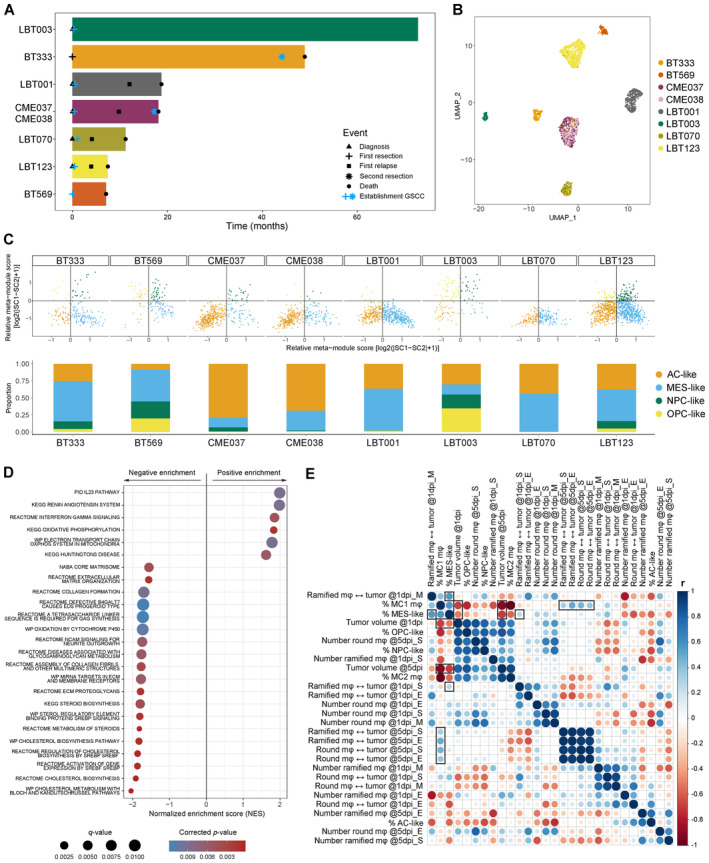
Figure 7 Macrophage/GAM‐GSCC interactions correlate to clinical outcome in GBM patients
Swimmer plot of included patients with indication of important events. UMAP plot of GBM tumor cells annotated by sample name. Two‐dimensional butterfly plot visualization of molecular subtype signature scores according to Neftel Dot plot showing the 25 most significant positive and negative enriched GSEA pathways using all curated gene sets of WikiPathways, Reactome, KEGG, PID and BioCarta databases for the group of invasive GSCCs (cut‐off corrected Correlogram of