- Title
-
Wnt signaling regulates neural plate patterning in distinct temporal phases with dynamic transcriptional outputs
- Authors
- Green, D.G., Whitener, A.E., Mohanty, S., Mistretta, B., Gunaratne, P., Yeh, A.T., Lekven, A.C.
- Source
- Full text @ Dev. Biol.
|
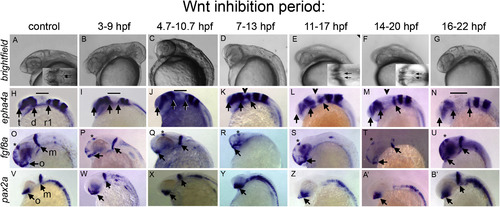
Timed inhibition of Wnt/β-catenin signaling reveals distinct neural plate responses. Embryos derived from hs:dkk1b/+ outcrosses were heat shocked to inhibit Wnt signaling during the time periods indicated above each column and in Fig. S2. All images: heads of 27 hpf embryos, anterior left, dorsal up. Brightfield (A–G). Insets in (A,E,F) are dorsal views, arrows indicate inner edge of MHB constrictions, which touch in control embryos. In situ hybridizations to epha4a (H–N), fgf8a (O–U), and pax2a (V–B’). Wild-type siblings to hs:dkk1b/+ embryos are shown in control (A,H,O,V). Note that Wnt inhibition between 3-9 hpf produces a range of dorsalized phenotypes; C3 dorsalized embryos are shown in (B,I,P,W). In situ results are equivalent in all dorsalized classes. (H–N) Arrows indicate telencephalon (t), diencephalon (d), rhombomere 1 (r1). Bar indicates midbrain, MHB and cerebellar primordia that do not express epha4a, indicated with arrowheads in (K,L,M). (O–B’) Arrows indicate optic stalk (o), and mhb (m, when present). Asterisks in (O–U) indicate dorsal diencephalon domain. |
|
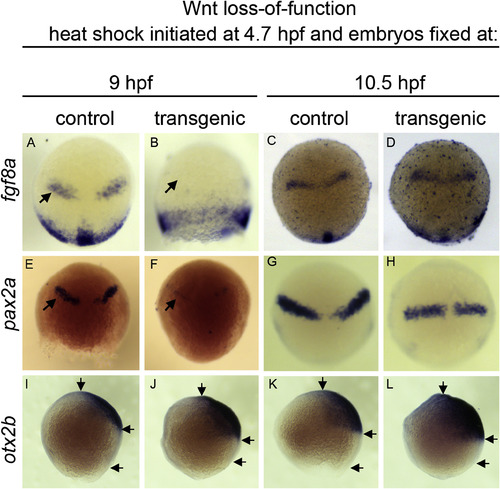
Wnt suppression transiently disrupts MHB establishment during epiboly. In situ hybridization to fgf8a (A–D), pax2a (E–H), and otx2b (I–L). hs:dkk1b/+ or +/+ sibling (control) embryos heat shocked at 4.7 hpf, then analyzed at 9 hpf (A,B,E,F,I,J) or 10.5hpf (C,D,G,H,K,L). (A–H) Dorsal view, animal pole up. (I–L) Lateral view, animal pole up. Note the absence of MHB associated fgf8a and pax2a at 9 hpf (B,F, arrows) but return of expression by 10.5 hpf (D,H). Arrows in (I–L) indicate anterior limit of otx2b, posterior limit of otx2b, and posterior edge of embryonic margin. Note shortened distance between the otx2b posterior limit relative to the margin in conditions of Wnt loss of function. |
|
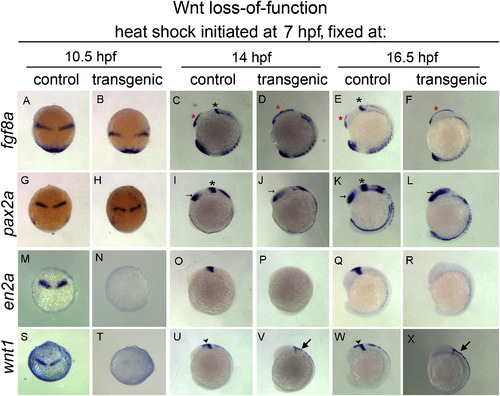
Wnt suppression during late epiboly/early segmentation leads to progressive loss of mes/r1 and expansion of forebrain. In situ hybridization to fgf8a (A–F), pax2a (G–L), en2a (M–R), and wnt1 (S–X) on hs:dkk1b/+ or +/+ sibling embryos. Dorsal views, anterior up (A,B,G,H,M,N,S,T). Lateral views, dorsal right, anterior up (C–F, I-L, O-R, U-X). Note normal fgf8a and pax2a at 10.5 hpf, despite significant reduction of wnt1 and en2a. By 14 hpf, MHB expression of fgf8 and pax2a is lost (indicated by black asterisks, C,I,E,K absent in D,J,F,L) with posterior expansion of telencephalic fgf8a (red asterisks, C–F) and optic stalk pax2a (arrows, I-L). At 16 hpf, posterior expansion of forebrain has increased (E,F,K,L). Midbrain and MHB wnt1 and en2a expression is absent, while dorsal hindbrain and spinal cord wnt1 persists (large arrows, V,X). . |
|
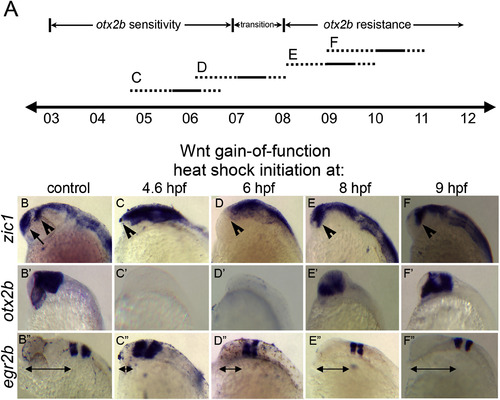
Timed global Wnt/β-catenin activation reveals two distinct windows of Wnt response. (A) Timeline (hours post fertilization) indicating periods of hs:wnt8a activation. Dashed lines represent uncertainty of beginning and end of Wnt pathway activation after heat shock. C,D,E,F: heat shock activation periods corresponding to images in bottom half of figure. (B–F”) Lateral views of heads of 2427 hpf embryos, anterior left. (B–F) In situ hybridization to zic1. (B’-F’) otx2b. (B”-F”) egr2b. Control: +/+ siblings of hs:wnt8a/+ embryos. Note absence of telencephalic zic1 domain (B, arrow) in all hs:wnt8a/+ embryos. Diencephalic zic1 domain is absent after 4.6 and 6 hpf heat shocks (B,E,F, arrowheads). otx2b expression is suppressed after 4.6 and 6 hpf heat shocks but is present, but shifted anteriorly, after 8 hpf and 9 hpf heat shocks (C’-F’). egr2b expression within r3 and r5 are shifted anteriorly in all treatments with the degree of anterior shift decreasing with later heat shocks (B”-F”, arrows). |
|
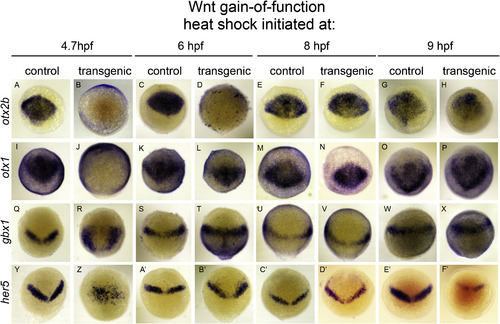
The neural plate responds dynamically to global Wnt activation during gastrulation. In situ hybridizations to otx2b (A–H), otx1 (I–P), gbx1 (Q–X), and her5 (Y–F’) on bud stage hs:wnt8a/+ and +/+ sibling (control) embryos. Dorsal views, animal pole up. Time of heat shock initiation is indicated above respective columns. Note repression of otx2 (B) and otx1 (J) after 4.7 hpf heat shock, concomitant with anterior expansion of gbx1 (R) and disorganized her5+ cells (Z). otx2b repression occurs after 6 hpf heat shock (D), though otx1 does not (L), gbx1 is mildly increased in the posterior neural plate (T), and her5 shows scattered expression anterior to normal domain (B’). Expression of all markers is relatively normal after 8 hpf heat shocks (F,N,V) with slight anterior expansion of her5 into prospective telencephalic domain (D’). otx2b, otx1 and gbx1 are normal after 9 hpf heat shock (H,P,X), while her5 is reduced (F’). |
|
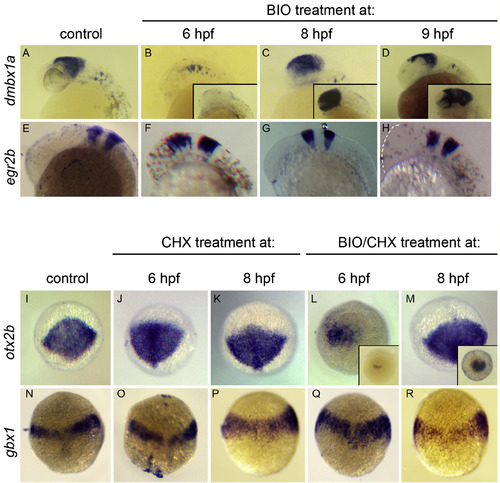
otx2b is directly suppressed by Wnt signaling. (A–H) BIO treatment recapitulates hs:wnt8a effects. In situ hybridization to dmbx1a in midbrain (A–D), egr2b in hindbrain (E–H). Insets in B,C,D show otx2b staining. 27hpf embryos anterior left, dorsal up. Note suppression of midbrain dmbx1a and otx2b after BIO treatment at 6 hpf (B) that is not observed with 8 or 9 hpf treatment (C,D). egr2b is shifted anteriorly, with the degree of anterior shift correlated with time of treatment (E-H; dotted line indicates anterior edge of embryo in H). (I–R) BIO treatment suppresses otx2b in absence of new protein synthesis. Analysis of 10 hpf embryos, oblique dorsal views, animal pole up, in situ hybridizations to otx2b (I–M) or gbx1 (N–R). (J,K,O,P) CHX treatment only. (L,M,Q,R) BIO + CHX treatment. Note the lack of neural plate otx2b after treatment with BIO + CHX at 6 hpf (L), similar to treatment with BIO alone (inset in L), which is not observed after treatment at 8 hpf (M; inset: BIO alone). Remaining mesodermal otx2b positive cells are visible, though numbers vary between embryos. gbx1 is not significantly affected with either CHX or CHX/BIO treatment (N–R). |
|
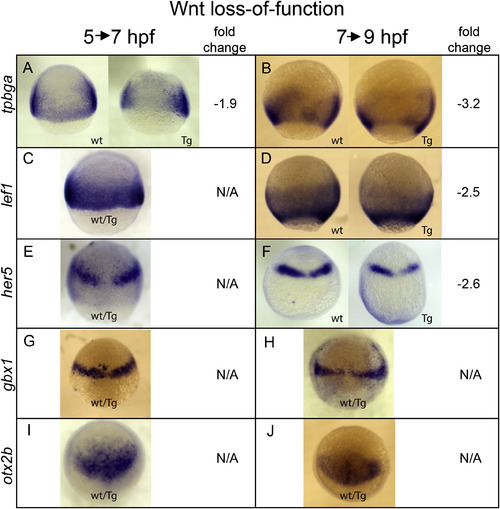
Validation of hs:dkk1b RNA-Seq analysis. Embryos collected from a hs:dkk1b/+ X +/+ cross were heat shocked during gastrulation according to experimental regimens outlined in Fig. S5 and fixed at 7 hpf (5→7 hpf experiment) or 9 hpf (7→9 hpf experiment) for in situ hybridization. Gene analyzed is indicated on left. Fold change is from the RNA-seq analysis (Supplemental Tables 1 and 2). wt: +/+ sibling. Tg: hs:dkk1b/+ embryo. wt/Tg: representative embryo from collection, genotype cannot be determined by staining pattern. Animal pole up, lateral views (A–D) or dorsal views (E–J). |
|
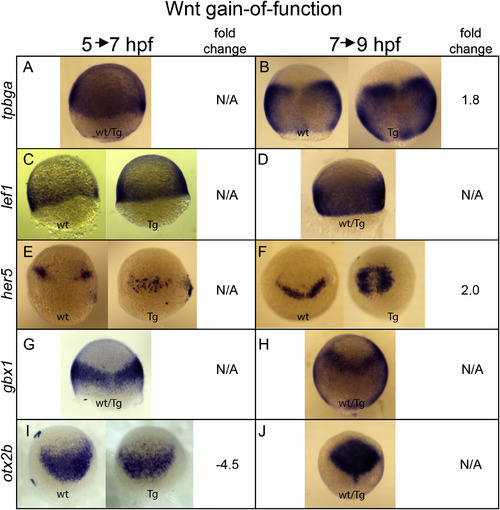
Validation of hs:wnt8a RNA-Seq analysis. Embryos collected from a hs:wnt8a/+ X +/+ cross were heat shocked during gastrulation according to experimental regimens outlined in Fig. S5 and fixed at 7 hpf (5→7 hpf experiment) or 9 hpf (7→9 hpf experiment) for in situ hybridization. Gene analyzed is indicated on left. Fold change is from the RNA-seq analysis (Supplemental Tables 1 and 2). wt: +/+ sibling. Tg: hs:dkk1b/+ embryo. wt/Tg: representative embryo from collection, genotype cannot be determined by staining pattern. Animal pole up, lateral views (A,C,D) or dorsal views (B,E-J). |
|
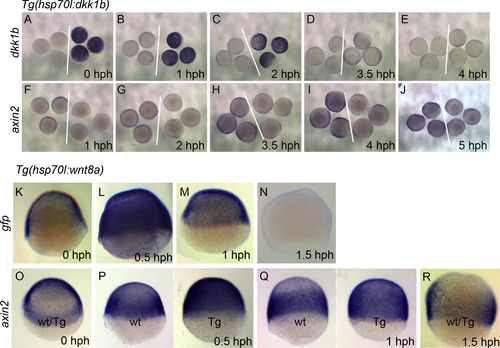
Temporal analysis of Wnt signaling modulation by the hs:dkk1b and hs:wnt8a transgenes. In situ hybridizations to dkk1b (A-E) or axin2 (F-J) in hs:dkk1b. Time of fixation after heat shock (hours post heat shock, hph) is indicated in the lower right. White lines separate wild type embryos (left of line) from transgenic embryos (right of line). dkk1b staining is strong up to 2 hph (A-C), declines by 3.5 hph (D), and is similar to wild type by 4 hph (E). axin2 is visibly reduced up to 3.5 hph (F-H), and recovers to normal levels between 4 hph and 5 hph (I,J). Note that the reduced staining in I is due to a reduction in neural plate staining that is a consequence of mispatterning, rather than a direct effect of the hs:dkk1b transgene. In situ hybridization to gfp (K-N) or axin2 (O-R) in hs:wnt8a. gfp staining is detectable 0.5 hph (L), is significantly reduced at 1 hph (M) and is not detectable by 1.5 hph (N). (O) axin2 staining is similar in hs:wnt8a embryos and wild-type siblings. (wt/Tg: genotype of embryos cannot be distinguished by staining). (P) axin2 is strongly induced at 0.5hph (Tg: hs:wnt8a embryo, wt: wild-type sibling; representative embryos shown). (Q) axin2 remains elevated at 1 hph. (R) axin2 levels are indistinguishable between hs:wnt8a and wild-type siblings. |
|
Schematic summary of hs:dkk1b treatments. Timeline of zebrafish development in hours post fertilization, with gastrulation and segmentation periods indicated for reference. Horizontal lines indicate periods of hs:dkk1b mediated Wnt knockdown. Dashed ends on each line indicate uncertainty regarding the beginning and end of active Wnt suppression during heat shock treatments. Based on the loss-of-function phenotypes shown in Fig. 1, we propose three phases of Wnt-mediated patterning; primary AP patterning, mes/R1, and MHB morphogenesis, as indicated above the time line. Arrows point out the heat shock periods shown in Fig. 1 |
|
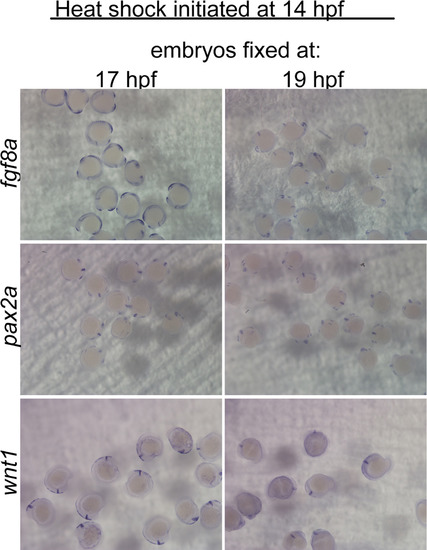
Wnt suppression during morphogenesis phase does not inhibit the MHB GRN before 19 hpf. In situ hybridizations for fgf8a, pax2a, and wnt1 on embryos from hs:dkk1b/+ X +/+ crosses, heat shocked at 14 hpf, and analyzed at the indicated times. No clear differences in gene expression between hs:dkk1b/+ and +/+ embryos could be observed at either 17 or 19 hpf. |
|
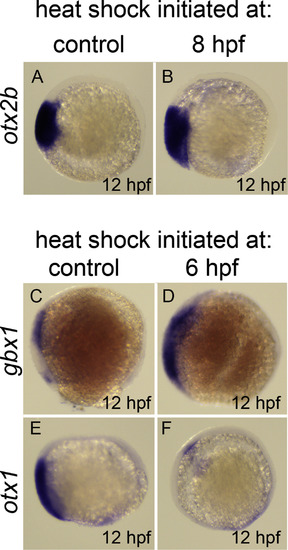
Dynamics of otx2b, otx1, and gbx1 response to hs:wnt8a. In situ hybridization of otx2b (A,B), gbx1 (C,D), and otx1 (E,F). 12 hpf embryos, lateral view, anterior left, dorsal up. (A,B) otx2b is not repressed by Wnt activation 4 hours post heat shock. (C,D) gbx1 expansion into anterior neural plate is observed 6 hours after induction of hs:wnt8a. This phenotype correlates with the absence of both otx1 expression (E,F) and otx2b (see Fig. 5D). |
|
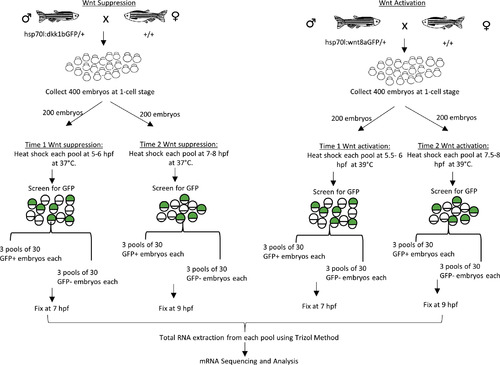
Schematic outline of embryo production procedure for gastrula stage Wnt modulation RNA-Seq analysis. hs:dkk1b/+ and hs:wnt8a/+ transgenic fish were crossed to WT strains. Embryos were collected and heat shocked two time points, 5/5.5 hpf (time 1) or 7/7.5 hpf (time 2) for hs:dkk1 and hs:wnt8a respectively. Embryos were sorted into GFP+ (transgenic embryos) and GFP- (wild-type sibling embryos) pools and homogenized in Trizol at 7 hpf (time 1) or 9 hpf (time 2). Total RNA was isolated and submitted to the Texas A&M AgriLife Research Genomics and Bioinformatics Service for library preparation and Illumina sequencing (See Material and Methods for more details). |
Reprinted from Developmental Biology, 462(2), Green, D.G., Whitener, A.E., Mohanty, S., Mistretta, B., Gunaratne, P., Yeh, A.T., Lekven, A.C., Wnt signaling regulates neural plate patterning in distinct temporal phases with dynamic transcriptional outputs, 152-164, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.