- Title
-
Identification of a novel interaction of FUS and syntaphilin may explain synaptic and mitochondrial abnormalities caused by ALS mutations
- Authors
- Salam, S., Tacconelli, S., Smith, B.N., Mitchell, J.C., Glennon, E., Nikolaou, N., Houart, C., Vance, C.
- Source
- Full text @ Sci. Rep.
|
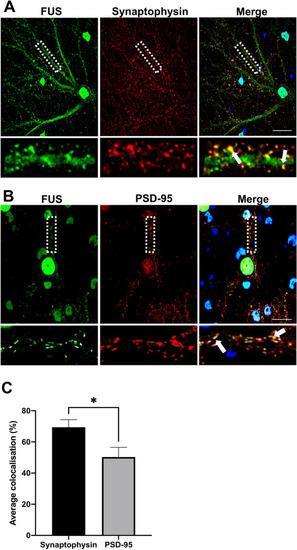
Subcellular localisation of FUS in primary cortical neurons. Immunofluorescent staining of DIV21 rat primary cortical neurons. ( |
|
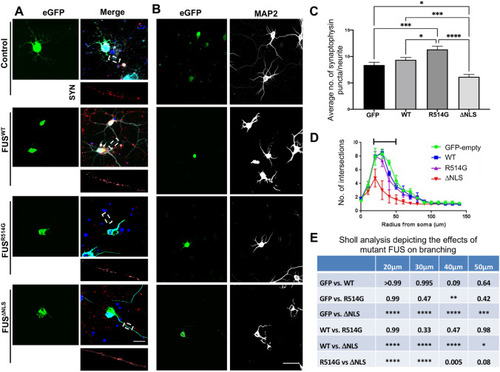
ALS-linked mutations in FUS lead to pre-synaptic alterations. ( |
|
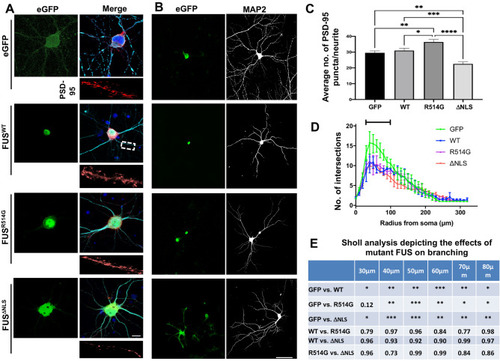
ALS-linked mutations in FUS lead to post synaptic alterations. Representative confocal images of DIV21 rat primary neurons transduced with eGFP, HA-FUSWT, HA-FUSR514G and HA-FUSΔNLS. Cells were stained for HA (green), PSD-95 (red) and MAP2 (merge). HA staining shows cytoplasmic mislocalisation of the mutant FUS. Regions of interest show magnified dendrites used to quantify the PSD-95. Nuclei were counterstained with DAPI. Scale bar = 10 μm. ( |
|
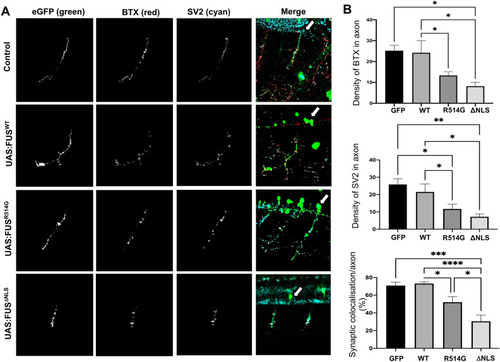
Zebrafish expressing mutant FUS show abnormal neuromuscular junctions and orphaned pre-synaptic endings. ( |
|
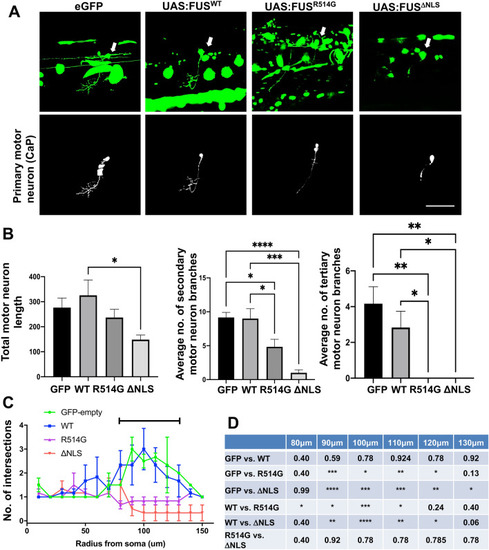
Mutant FUS expression in primary motor neurons affects axonal branching. ( |
|
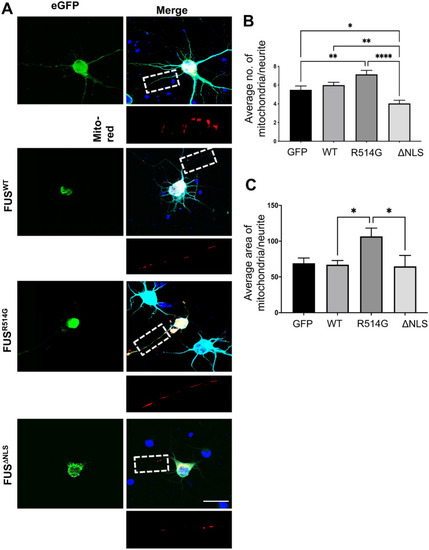
Expression of mutant FUS leads to mitochondrial abnormalities. ( |
|
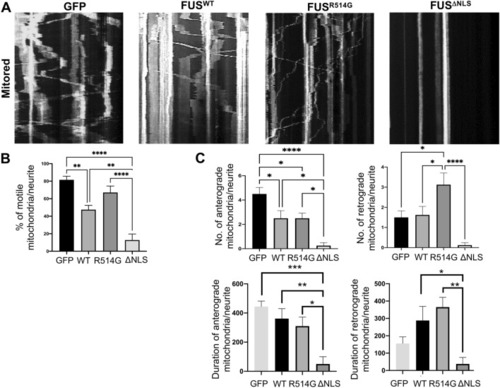
Mitochondrial motility is affected by mutations in FUS. ( |
|
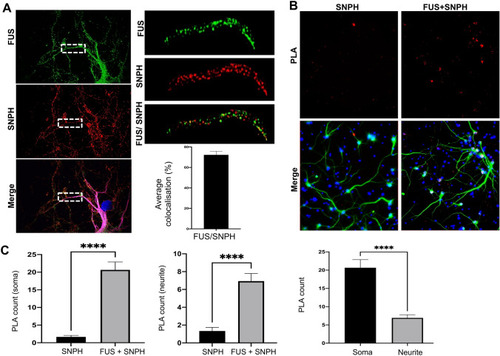
FUS Interacts with the mitochondrial anchor protein, Syntaphillin. ( |
|
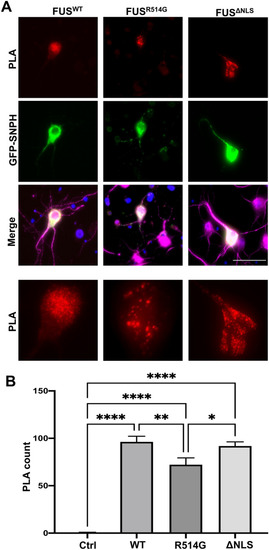
Overexpression of FUS mutations and GFP-SNPH alters FUS-SNPH interactions in transfected neurons. ( |
|
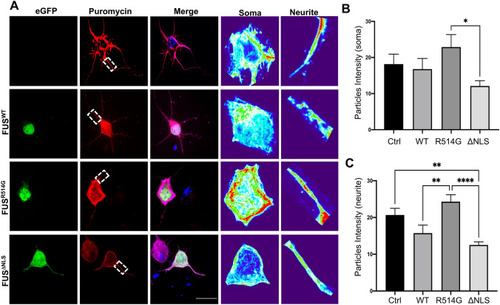
Overexpression of FUS mutations alters protein translation within transfected neurons. ( |