Figure 10.
- ID
- ZDB-FIG-231124-28
- Publication
- Celotto et al., 2023 - Single-cell RNA sequencing unravels the transcriptional network underlying zebrafish retina regeneration
- Other Figures
-
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 2.
- Figure 1—figure supplement 3.
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 2.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 2.
- Figure 4.
- Figure 5.
- Figure 6.
- Figure 7.
- Figure 8—figure supplement 1.
- Figure 8—figure supplement 1.
- Figure 8—figure supplement 2.
- Figure 9—figure supplement 1.
- Figure 9—figure supplement 1.
- Figure 10.
- All Figure Page
- Back to All Figure Page
|
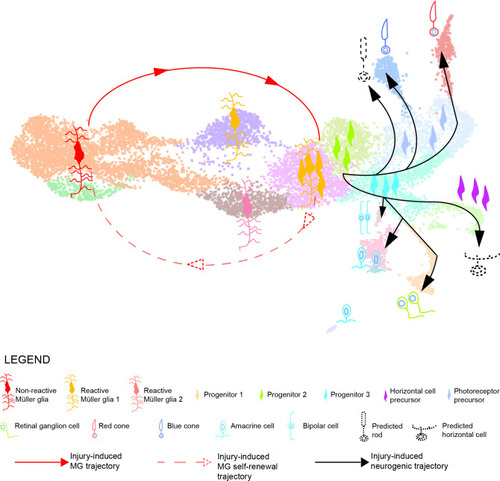
Model summarizing the cellular events underlying retina regeneration in zebrafish. Upon lesion, non-reactive MG on the left side of the main transcriptional UMAP feed into a hybrid Müller glia/progenitor population via reactive Müller glia (solid red line). Based from this hybrid cell population, we find a Müller glia trajectory reverting to the initial starting point indicating Müller glia self-renewal (dashed red line). Moreover, the hybrid cell population represents the tip of the neurogenic trajectory, resulting in the formation of neuronal precursors and differentiating neurons (solid black lines). Differentiating rods and horizontal cells, which have not been found in the scRNAseq, are indicated with dashed lines and positioned in the predicted location on the main UMAP. |