- Title
-
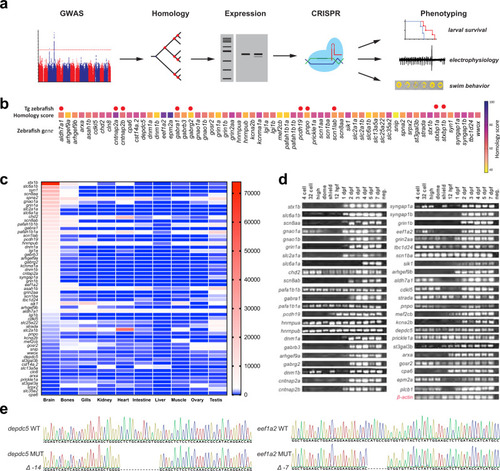
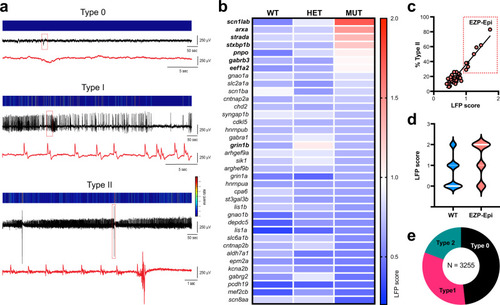
Phenotypic analysis of catastrophic childhood epilepsy genes
- Authors
- Griffin, A., Carpenter, C., Liu, J., Paterno, R., Grone, B., Hamling, K., Moog, M., Dinday, M.T., Figueroa, F., Anvar, M., Ononuju, C., Qu, T., Baraban, S.C.
- Source
- Full text @ Commun Biol
|
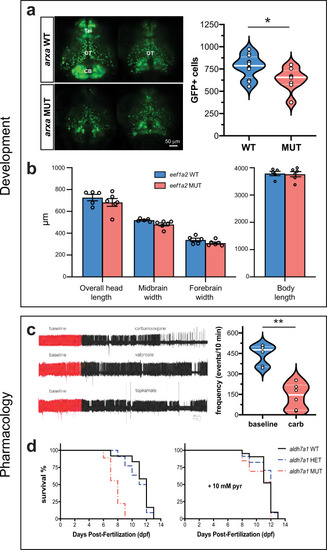
EXPRESSION / LABELING:
|
|
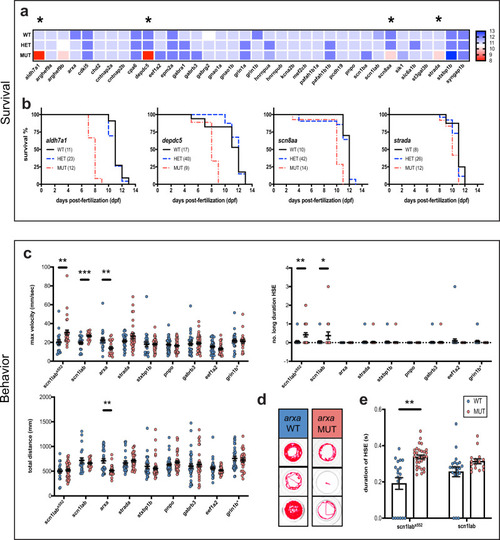
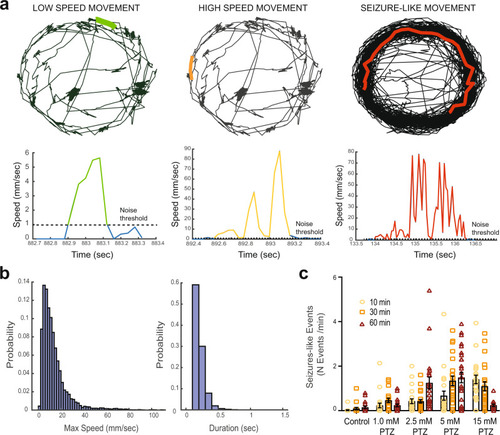
PHENOTYPE:
|
|
|
|
|
|
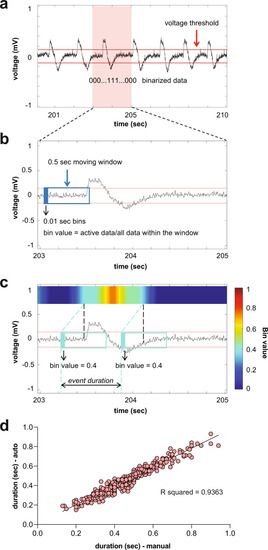
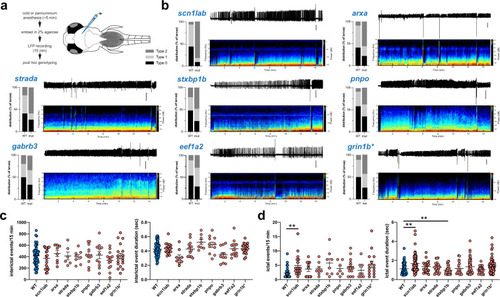
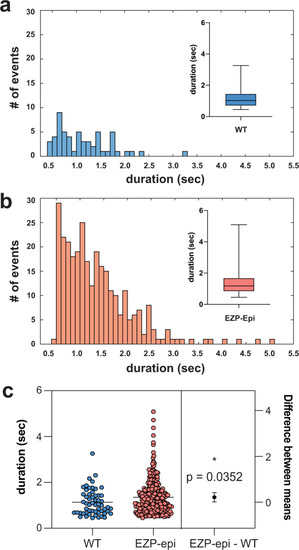
Histograms depict number and duration of ictal events measured using a custom MATLAB-based program for PHENOTYPE:
|
|
|
|
|
|
|