- Title
-
Pbx acts with Hand2 in early myocardial differentiation
- Authors
- Maves, L., Tyler, A., Moens, C.B., and Tapscott, S.J.
- Source
- Full text @ Dev. Biol.
|
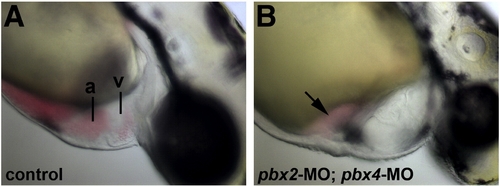
Defective heart development in pbx2-MO; pbx4-MO embryos. (A–B) Lateral views of live (A) wild-type and (B) pbx2-MO; pbx4-MO embryos at 48 hpf. pbx2-MO; pbx4-MO embryos show mild pericardial edema and blood pooled near the atrium (arrow). Anterior is to the right. a, atrium. v, ventricle. PHENOTYPE:
|
|
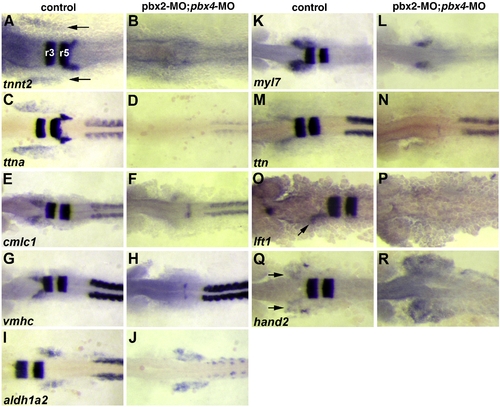
Validation of microarray-identified Pbx-dependent heart genes. (A–R) RNA in situ expression of genes from Table 1 in (A, C, E, G, I, K, M, O, Q) wild-type control or (B, D, F, H, J, L, N, P, R) pbx2-MO; pbx4-MO embryos. (A–D) are at 10 somite stage (10s). (E–N,Q–R) are at 18s. (O–P) are at 20s to achieve more robust expression of lft1 in controls. krox-20, labeled in hindbrain rhombomeres r3 and r5 in (A), is included in all in situs to control for loss of Pbx. The wild-type heart primordium expression domains of tnnt2, ttna, cmlc1, vmhc, myl7, lft1 and hand2 are just anterior-lateral to r3 krox-20 expression (arrows in A, O, Q). n ≥ 10 for each marker in control or pbx2-MO; pbx4-MO. Embryos are shown in dorsal view, anterior towards the left. EXPRESSION / LABELING:
|
|
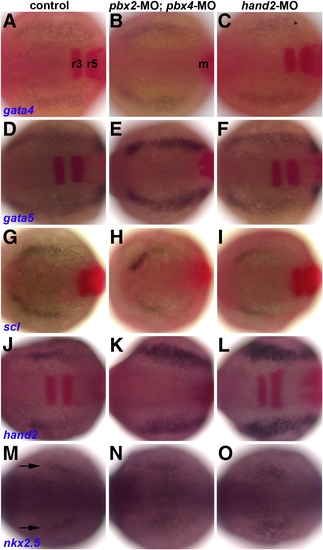
Early heart specification is normal in pbx2-MO; pbx4-MO and hand2-MO embryos. (A–O) RNA in situ expression at the 7–9 somite stage of (A–C) gata4, (D–F) gata5, (G–I) scl, (J–L) hand2, and (M–O) nkx2.5, all in blue, in (A, D, G, J, M) control, (B, E, H, K, N) pbx2-MO; pbx4-MO, or (C, F, I, L, O) hand2-MO embryos. Also included in (A–L), in red, are krox-20, labeled in (A) in r3 and r5 in the hindbrain, and myod, labeled m in (B) and used to confirm somite staging, but is not visible here in all embryos. Arrows in (M) mark nkx2.5 expression. n ≥ 13 for each marker in control, pbx2-MO; pbx4-MO, or hand2-MO. Embryos are shown in dorsal view, anterior towards the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
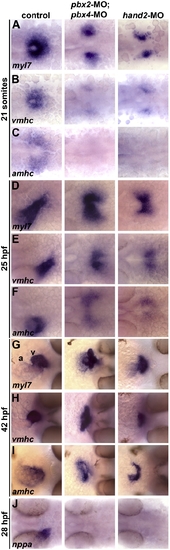
Pbx-deficient and hand2-MO embryos exhibit similar cardiac differentiation and morphogenesis defects. (A–J) RNA in situ expression of (A,D,G) myl7, (B,E,H) vmhc, (C,F,I) amhc, and (J) nppa at (A–C) 21 somites, (D–F) 25 h post fertilization (hpf), (G–I) 42 hpf, and (J) 28 hpf. In control embryos shown, myl7 labels all myocardial precursors, vmhc labels ventricular precursors, and amhc labels atrial precursors (Berdougo et al., 2003). Ventricle (v) and atrium (a) are labeled in (G). n ≥ 20 for each marker in control, pbx2-MO; pbx4-MO, or hand2-MO. (A–F) Embryos are shown in dorsal view, anterior towards the left. (G–I) Embryos are shown in frontal views, dorsal towards the right. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Pbx and Hand2 genetically interact in cardiac morphogenesis. (A–E) RNA in situ expression of myl7 at 42 hpf in (A) control, (B) pbx2-MO; pbx4-MO, (C) hand2-MO, (D) pbx2-MO; pbx4-MO; hand2-MO, or (E) hand2-/- embryos. Embryos are shown in frontal views, dorsal towards the right. (F–G) Quantification of phenotypes observed using myl7 expression at (F) 24 hpf or (G) 42 hpf. Normal phenotypic classes (black) correspond to tube (24 hpf) or looped (42 hpf) patterns depicted in Fig. 4D and subpanel A of this figure, respectively. Delayed or abnormal phenotypic classes (gray) correspond to images shown for pbx2-MO; pbx4-MO and hand2-MO embryos in Fig. 4D (24 hpf) or subpanels B–C of this figure (42 hpf). The separate, or cardia bifida, phenotypic classes (white) correspond to two myl7 domains and resemble those described for pbx2-MO; pbx4-MO and hand2-MO embryos in Fig. 4A or for pbx2-MO; pbx4-MO; hand2-MO and hand2-/- embryos in subpanels D–E of this figure. Numbers in parentheses denote numbers of embryos. hand2-/- embryos were derived from clutches that contained about 25% mutant embryos; their hand2+/+ and +/- siblings showed normal myl7 expression. |
|
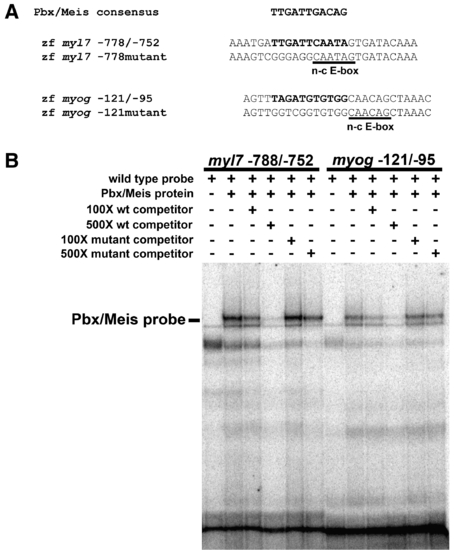
Pbx proteins bind the myl7 promoter in vitro. (A) Pbx/Meis consensus binding site sequence (Berkes et al., 2004) aligned relative to putative Pbx binding site sequences (highlighted in bold) in the zebrafish myl7 promoter and myogenin (myog) promoter. The non-canonical E-box (n-c E-box) sites are underlined. Also shown are sequences of mutated Pbx binding sites. Sequences represent oligos used in EMSA. (B) Pbx4 and Meis3 proteins were generated by in vitro translation using a reticulocyte lysate and subjected to EMSA with the specified myl7 or myog probes. Pbx/Meis-bound probe band is indicated. The non-specific bands were observed in multiple gel shifts. Addition of unlabeled myl7 -778/-752 probe or myog -121/-95 probe decreases binding of Pbx/Meis, whereas addition of mutant unlabeled probes does not. |
|
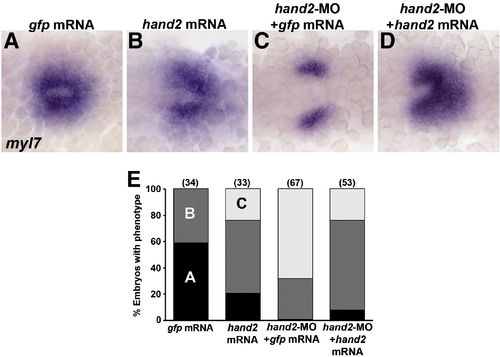
hand2 RNA rescues myl7 expression in hand2-MO embryos. (A–D) RNA in situ expression of myl7 at the 20s stage in embryos injected with (A) control gfp mRNA, (B) hand2 mRNA, (C), hand2-MO and gfp mRNA, or (D) hand2-MO and hand2 mRNA. Embryos are shown in dorsal view, anterior towards the left. (A) In 20s control embryos, heart precursors form a cone on the embryonic midline. (B) Embryos injected with hand2 mRNA have about wild-type numbers of myl7-expressing cells but heart morphogenesis and cone formation is slightly impaired. (C) Embryos injected with hand2-MO have reduced lateral myl7 domains with defective midline migration. (D) hand2 mRNA largely rescues the hand2-MO phenotype, although most embryos show some impaired morphogenesis. (E) Quantification of the rescue data. Numbers in parentheses denote numbers of embryos. The phenotypic classes correspond to the images shown in panels A–C. |
Reprinted from Developmental Biology, 333(2), Maves, L., Tyler, A., Moens, C.B., and Tapscott, S.J., Pbx acts with Hand2 in early myocardial differentiation, 409-418, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.