- Title
-
Innate immune evasion revealed in a colorectal zebrafish xenograft model
- Authors
- Póvoa, V., Rebelo de Almeida, C., Maia-Gil, M., Sobral, D., Domingues, M., Martinez-Lopez, M., de Almeida Fuzeta, M., Silva, C., Grosso, A.R., Fior, R.
- Source
- Full text @ Nat. Commun.
|
|
|
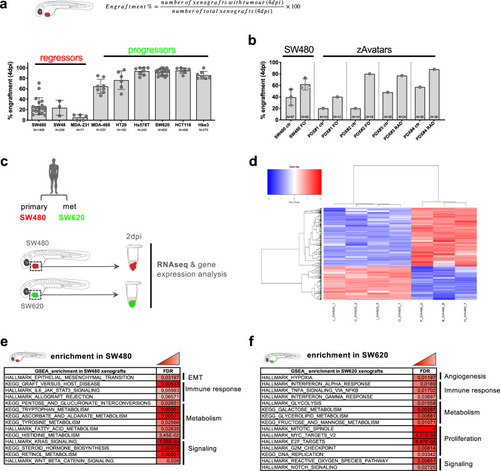
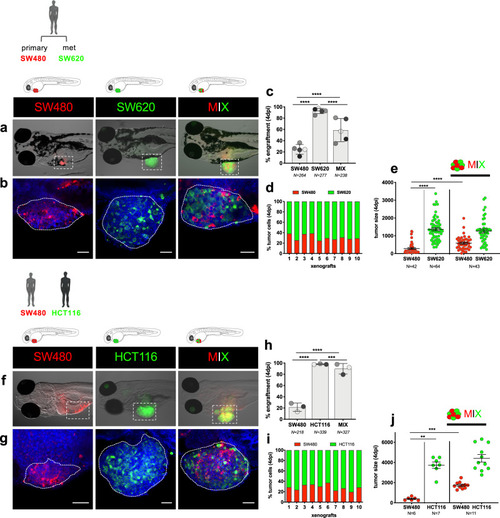
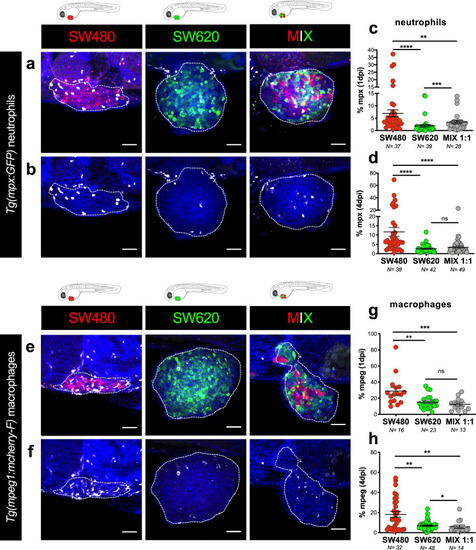
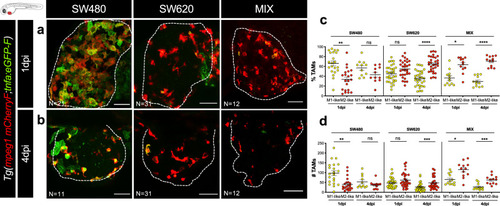
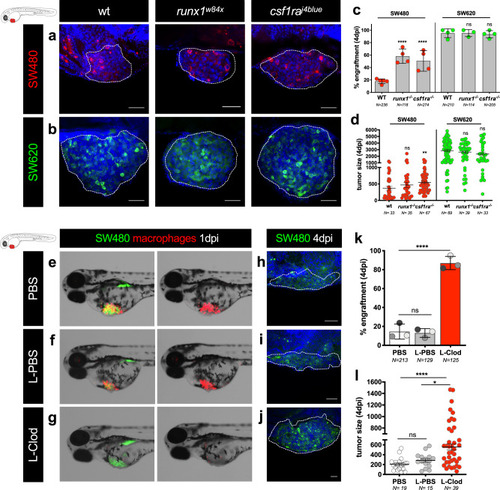
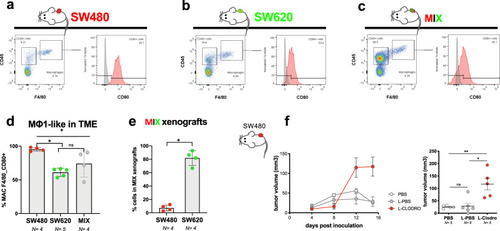
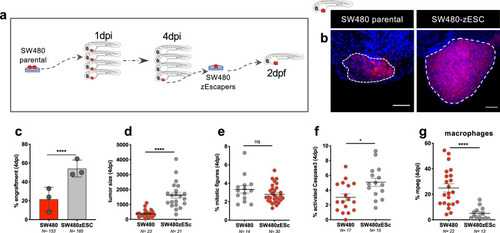
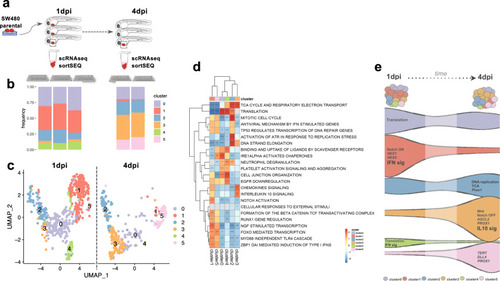
Tumor cells were labeled with lipophilic dyes and injected into the PVS of 2dpf zebrafish embryos. |
|
|
|
|
|
|
|
|
|
|
|
|