- Title
-
Automated whole animal bio-imaging assay for human cancer dissemination
- Authors
- Ghotra, V.P., He, S., de Bont, H., van der Ent, W., Spaink, H.P., van de Water, B., Snaar-Jagalska, B.E., and Danen, E.H.
- Source
- Full text @ PLoS One
|
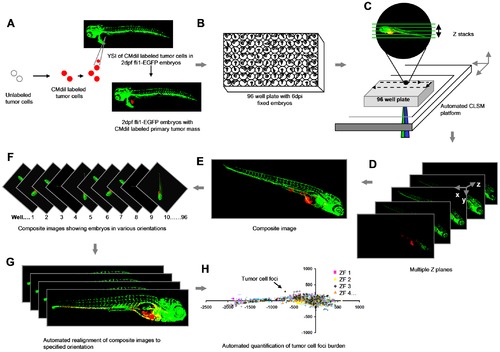
Schematic overview of the procedure. (A) Yolk sac implantation of CM-DiI labeled tumor cells into Tg (Fli:EGFP) ZF embryos 2 days post-fertilization. (B) Formaldehyde fixed 6 dpi embryos arrayed in 96 well plates. (C) Automated image acquisition using CLSM platform equipped with movable stage captures multiple Z stacks per embryo using 488 and 561 nm laser lines. (D and E) Automated creation of extended depth composite images. (F) Multiple extended depth images depicting embryos lying in different lateral orientations. (G) Automated uniform reorientation of images. (H) Scatter plot representing tumor foci burden in multiple embryos belonging to one experimental condition. |
|
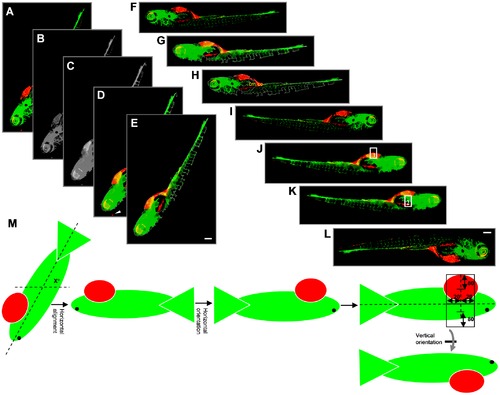
Outline of steps involved in embryo orientation. (A) Extended depth image of 6 dpi ZF embryo. (B) Grey value image from combination of green and red channels. (C) Blurred grey image after applying closing filter to optimize determination of outline. (D) Embryo segmented after applying intensity threshold and area filter. Arrowhead indicates a red object outside the outline that is excluded from segmentation. (E) Cropped image with only selected object. (F) Embryo rotated by x° for horizontal reorientation. (G and H) Determination of the x position value of the center of mass (cm) and center of centroid (cc). (I) Horizontal flip of the image only if cm is on the left side of cc, resulting in images with the head of the embryo always to the right side. (J) Image after applying closed filter to the combined green and red channel to get the outline of the embryo. Point lying at 75% distance from the extreme left of the embryo outline is calculated. Y-axis is drawn at this X-position from upper to lower outline. Upper rectangle 1 is drawn. (K) Lower rectangle 2 is drawn. (L) Vertical flip of the image only if red intensity in rectangle 1 is higher than in rectangle 2. (M) Schematic representation of calculations for steps E–I. Altogether, this procedure results in images where the head is on the right and the yolk sac is on the bottom of the image. Scale bar = 200 μm in E and I. |
|
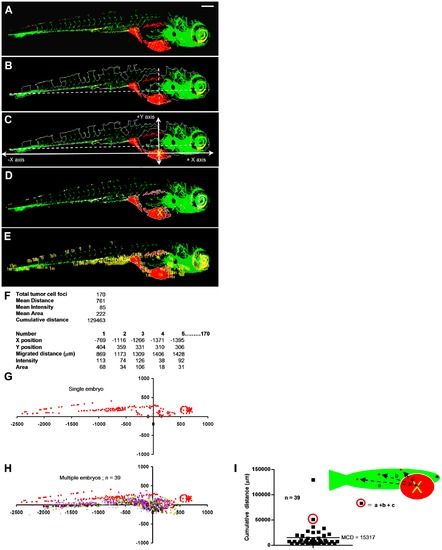
Automated multiparametric quantification of PC3 tumor foci. (A) Extended depth image of 6 dpi fixed embryo after realignment. (B) Embryo outline from segmented GFP channel and Y-axis intersecting X-axis at 75% from extreme left. (C) Calculated injection point at 75% distance from the extreme left and 75% from the top Y position. (D) Segmented red channel showing tumor foci burden in the embryo. (E) Identified tumor foci. (F) Multiple parameters of tumor foci burden calculated per embryo. Each number in the image corresponds to one tumor focus. (G) Tumor foci dissemination in a single embryo represented as scatter plot (coordinates 0,0 represents calculated injection site). (H) Combined scatter plot showing tumor foci dissemination from 39 injected embryos. (I) Quantification of cumulative distance (CD). Each filled square represents cumulative distance from injection point of all identified tumor foci in a single embryo. Mean cumulative distance (MCD) in the 39 injected embryos in this experiment is 15024 μm. Scale bar = 200 μm in A. |
|
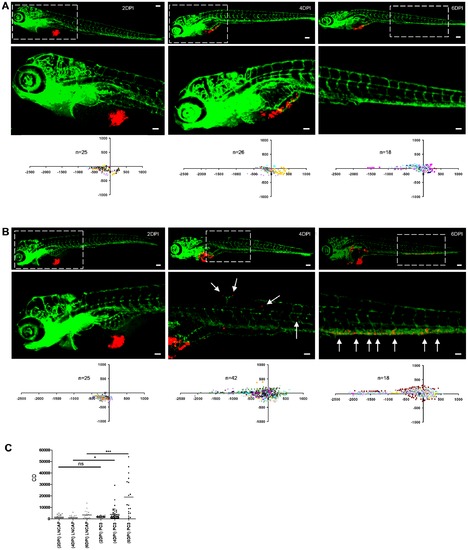
Determination of cancer cell dissemination kinetics. (A and B) LnCAP (A) or PC3 cells (B) were implanted and embryos were fixed at 2, 4, or 6 dpi for imaging (immunofluorescence images and automated image analysis (scatter plots)). Bottom row images (scale bar = 50 μm) show zoom-in of area marked by dotted line in top row images (scale bar = 100 μm). (C) CD at 2, 4 and 6 dpi for LnCAP (grey) and PC3-injected embryos (black) calculated from scatterplots in A and B, respectively. Statistical testing for difference between LnCAP and PC3 at different dpi is indicated. *p<0.05, ***p<0.001. |
|
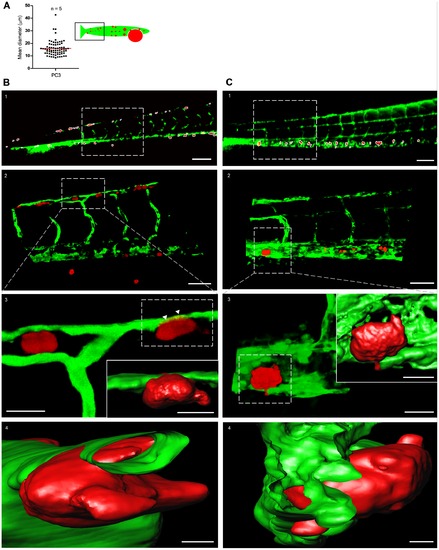
Characterization of tumor cell foci identified by macro using high-resolution imaging. (A) Quantification of the mean diameter of macro-identified tumor cell foci from the tail region of PC3-injected embryos. Data obtained from 5 embryos. (B) High resolution imaging of CM-DiI-labeled PC3 tumor cell foci. (B1) Macro-identified PC3 tumor cell foci. (B2) Zoom-in on area indicated in B1 shows tumor cells in association with host vasculature. (B3 and B4) Three dimensional reconstruction and surface rendering of area in insert of B2; arrowheads point to tumor cell partly inside distal longitudinal anastomotic vessel (Video S2 and S3). (C) High resolution imaging of PC3-mCherry tumor cell foci. C1–4, as B1–4 for PC3-mCherry. Scale bar is 100 µm in B1 and C1; 50 μm in B2 and C2; 15 μm in B3 and C3; 10 μm in insets in B3 and C3; 5 μm in B4 and C4. |
|
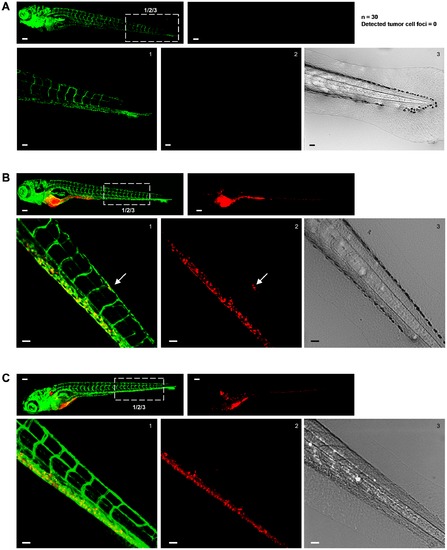
Excluding disturbance of the image analysis by autofluorescence signal from pigment cells. (A–C) In each case top left image shows green signal (Fli-EGFP) and top right image shows red signal for tumor cells. Bottom images show zooms of boxed area in top left image providing green (left) and red signal (middle) and transmitted light (right). Scale bar is 100 μm in images showing whole embryo and 50 μm in zoomed images. (A) Non-implanted fli-EGFP embryo imaged at 8 days post fertilization. Number of non-implanted embryos and number of tumor cells (falsely) detected by automated imaging and image analysis method is indicated at the right. (B) Fli-EGFP embryo implanted with CM-DiI-labeled PC3 imaged at 6 dpi. (C) Fli-EGFP Casper embryo implanted with CM-DiI-labeled PC3 imaged at 6 dpi. |
|
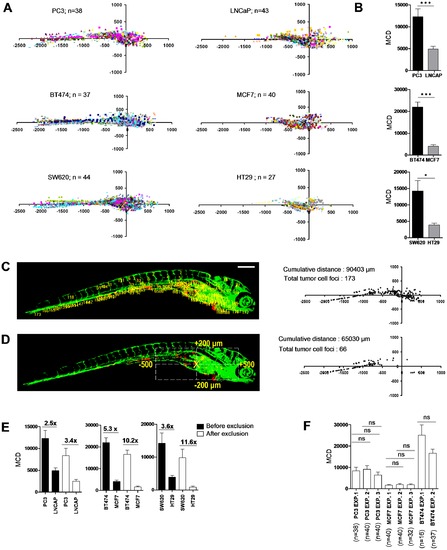
Differentiation between poorly and highly aggressive human cancer cell lines using automated bioimaging assay. (A) Scatter plot representation of tumor cell dissemination for indicated prostate (upper graphs), breast (middle), and colorectal cancer cell lines (lower graphs). Number of injected embryos from 2 biological replicates is indicated. (B) MCD determined from data represented in A. Data are presented as mean ± s.e.m. *p<0.05, ***p<0.001. (C) 6 dpi embryo injected with PC3 showing tumor foci burden determined from segmented red channel (left), and represented as scatter plot (right). (D) Automated determination of region for exclusion of tumor foci around implantation site and in area of intestinal development (left), and remaining tumor foci represented as scatter plot (right). (E) MCD before (black) and after exclusion (white bars) for the indicated prostate (left), breast (middle), and colorectal cancer lines (right graph). Fold difference between poorly and highly aggressive cell lines is indicated. Data are presented as mean ± s.e.m. *p<0.05, ***p<0.001. (F) MCD after exclusion for PC3 and MCF7 in multiple independent experiments demonstrates reproducibility. Data are presented as mean ± s.e.m. |
|
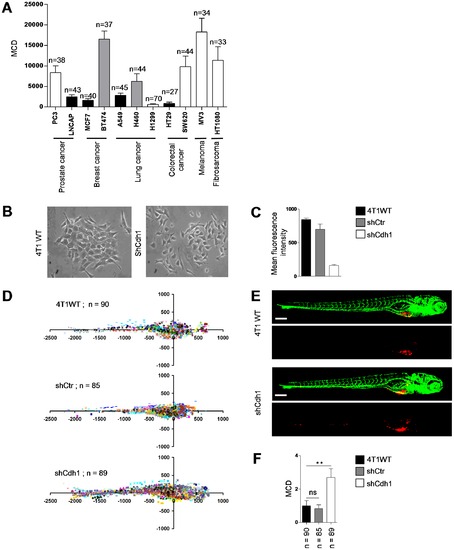
Differentiation between epithelial and mesenchymal cell types using the automated bioimaging assay. (A) MCD in a panel of human cancer cell lines from different origins. Number of injected embryos is indicated. White bars indicate cell lines showing a scattered phenotype in 2D cell culture. Black bars indicate cell lines growing as epithelial islands in 2D culture. Grey bars indicate cell lines with intermediate/mixed epithelial/mesenchymal characteristics. (B) 4T1 breast cancer cells growing as islands of loosely attached spindle-shaped cells (left) and completely scattered growth of 4T1 cells following E-cadherin silencing (right). (C) E-cadherin surface expression by FACS. (D) Scatter plot representation of indicated 4T1 variants. Number of injected embryos from 2 independent experiments is shown. (E) Representative images of embryos injected with indicated 4T1 variants. (F) MCD determined from data represented in D. Data are presented relative to wild type 4T1 as mean ± s.e.m. **p<0.01. Scale bar is 200 μm in E. |
|
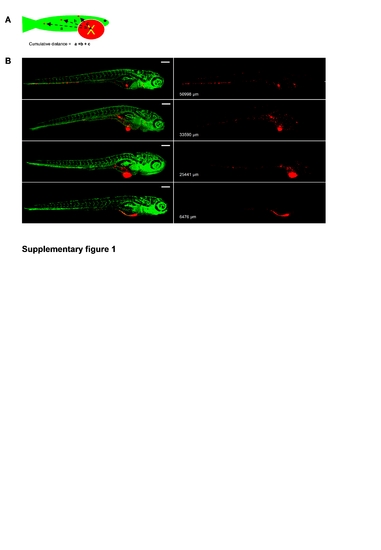
Automatically calculated cumulative distance (CD) in 6 dpi PC3 implanted embryos correlates with visual inspection of tumor cell dissemination. A, Scheme depicting concept of CD of tumor cell-foci. B, left images show CM-DiI-labeled tumor cells in red and GFP-endothelial cells of the Tg (Fli:GFP) line in green. Right images show only CM-DiI signal and calculated CD is indicated for each embryo. |
|
Representative image of BT474 (left) and MCF7 (right) implanted 6 dpi embryo. Scale bar is 200 μm. |