- Title
-
Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b
- Authors
- Rohrschneider, M.R., Elsen, G.E., and Prince, V.E.
- Source
- Full text @ Dev. Biol.
|
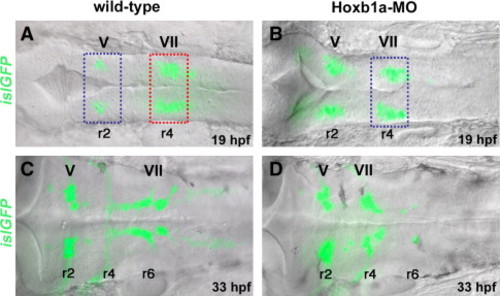
Microarray experimental strategy. islet1-GFP transgenic embryos in dorsal view, bright field (DIC optics) and fluorescent images merged. Anterior to the left in all panels. Rhombomeres (r) 2, r4, and r6 are labeled. (A, B) At 19–20 hpf, islet1-GFP-positive branchiomotor neurons are present in wild-type (A) and Hoxb1a-deficient embryos (B; injected with Hoxb1a-MO). Hoxb1a-positive tissue dissected from unmanipulated r4 (A, red box) was compared to Hoxb1a-deficient tissue dissected from r4 of Hoxb1a-MO injected embryos (B, blue box), and from unmanipulated r2 (A, blue box). (C, D) By 33 hpf, GFP-positive facial neurons have migrated into the posterior hindbrain of unmanipulated embryos (C), but remain in r4 of Hoxb1a-MO injected embryos (D). ov: otic vesicle; MO: morpholino; V: trigeminal neurons; VII: facial neurons. |
|
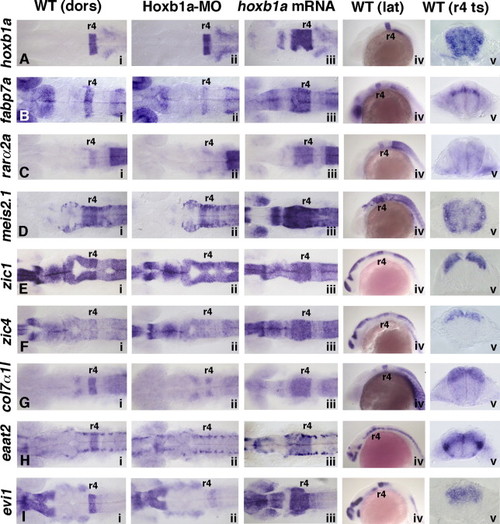
Expression of validated Hoxb1a target genes with broad r4 expression domains. In situ hybridization at 20 hpf of hoxb1a (A) and eight Hoxb1a target genes (B–I). Anterior to the left in all panels. Rhombomere (r) 4 is labeled. Panels i and iv: In wild-type embryos, (A) hoxb1a is expressed in an r4 stripe, as are (B) fabp7a, (C) rarα2a, (G) col7α1l, (H) eaat2, and (I) evi1. Expression of (D) meis2.1, (E) zic1, and (F) zic4 is elevated in r4 as compared to the surrounding hindbrain tissue. The eight Hoxb1a target genes also have Hoxb1a-independent expression domains, such as the lower-level (E) zic1 and (F) zic4 expression along the length of the neural tube, and (H) the bilateral discrete domains of eaat2. Panel ii: In Hoxb1a-deficient embryos, (A) hoxb1a mRNA remains unaffected, however, the r4-specific expression of all eight Hoxb1a target genes (B–I) is missing or reduced. Panel iii: hoxb1a mRNA-injected embryos show a characteristic anterior expansion of (A) hoxb1a and (B–I) all eight Hoxb1a target genes into r2 and r3 (and some more anterior regions). Panel v: Transverse sections through wild-type embryos show that (A) hoxb1a, (C) rarα2a, and (D) meis2.1 are expressed throughout the DV extent of the neural tube. (B, E–I) The remaining six genes are expressed in varying subdomains along the DV axis. dors: dorsal; lat: lateral; MO: morpholino; ts: transverse. EXPRESSION / LABELING:
PHENOTYPE:
|
|
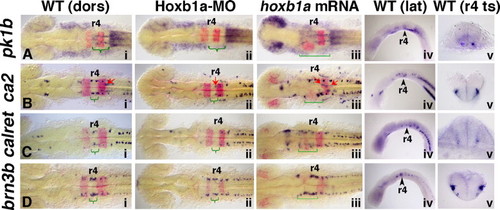
Expression of validated Hoxb1a target genes with neuronal specific expression domains within r4. In situ hybridization at 20 hpf demonstrates that pk1b, ca2, calret and brn3b are regulated by Hoxb1a. Anterior to the left in all panels. Rhombomere (r) 4 is labeled. krox20 marks r3 and r5 (panels i–iii; in red). Curved green brackets indicate Hoxb1a-dependent expression (compare panels i with ii). Square green brackets indicate expanded Hoxb1a-dependent expression in panel iii. Panels i and iv: (A) In wild-type embryos, pk1b is expressed in narrow bilateral trails through r5 and r6 (green bracket) as well as at low levels in ventral r4. (B) ca2 is expressed in bilateral discrete domains in r4, r5, and r6 in wild-type embryos (green bracket, Hoxb1a-dependent expression in r4), as well as in the trigeminal (r2) and facial (r5/6; red arrows) BMNs. (C) calret is expressed bilaterally in the hindbrain in three domains in wild-type embryos (green bracket, Hoxb1a-dependent expression in r4). (D) brn3b is expressed bilaterally in r3–r5 in wild-type embryos, with an elevated expression domain in r4 (green bracket, Hoxb1a-dependent expression). Panel ii: The r4-specific expression of all four genes is missing or reduced in Hoxb1a-MO-injected embryos (green brackets). Note that in panel Bii, while the four Hoxb1a-dependent domains are missing from r4, the Hoxb1a-independent ca2 expression is retained in the facial BMNs, which fail to migrate from r4 in Hoxb1a-deficient embryos (red arrow). Panel iii: The r4-specific expression of all four genes is anteriorly expanded in hoxb1a mRNA-injected embryos (square green brackets). Note that in panel Biii, the bilateral Hoxb1a-dependent ca2 expression is expanded anteriorly (square green brackets), and the Hoxb1a-independent expression of ca2 continues to mark the facial BMNs which have also expanded anteriorly (red arrows). Panel v: Transverse sections through wild-type embryos show that (A) pk1b is expressed in discrete bilateral ventral cells in r5 and at low levels throughout ventral r4 (section includes both r4 and r5). (B–D) Hoxb1a-dependent expression of ca2, calret, and brn3b is found laterally within the ventral half of the neural tube. dors: dorsal; lat: lateral; MO: morpholino; ts: transverse. EXPRESSION / LABELING:
PHENOTYPE:
|
|
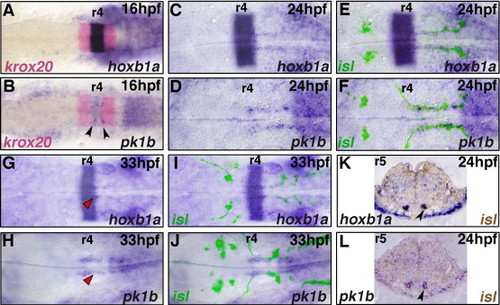
hoxb1a and pk1b are expressed in migrating facial BMNs. In situ hybridization for hoxb1a (A, C, E, G, I, K) and pk1b (B, D, F, H, J, L) during facial BMN migration (blue); krox20 marks r3 and r5 (A, B, red). islet1-GFP (isl) expressing BMNs were labeled with anti-GFP antibody (fluorescence in panels E, F, I, J; brown staining in panels K, L). Embryos are flat-mounted with anterior to the left (A–J), or transverse 5 μm plastic sections through r5 (K, L). Rhombomeres (r) 4 and r5 are labeled. At 16 hpf, (A) hoxb1a and (B) pk1b are expressed in a stripe in r4; arrowheads in (B) indicate elevated pk1b expression in discrete domains within r4. By 24 hpf, (D) pk1b is no longer expressed in a stripe in r4, however both (C) hoxb1a and (D) pk1b are expressed in narrow bilateral trails stretching from r4 to r7. (E, F) This expression co-localizes with the facial BMNs along the length of their migration (C, D, bright-field; E, F same images merged with fluorescent anti-GFP label). (K, L) Co-localization is confirmed by transverse sections through r5. Arrowheads indicate facial BMNs labeled by both islet1-GFP (brown) and (K) hoxb1a, or (L) pk1b. At 33 hpf, (G) hoxb1a and (H) pk1b expression is significantly reduced in the facial BMNs, and is present in narrow bilateral domains in r4 and r5 which flank the facial neuron axon tracts marked by islet1-GFP (red arrowheads) (G, H, bright-field; I,J, same images merged with fluorescent anti-GFP label). EXPRESSION / LABELING:
|
|
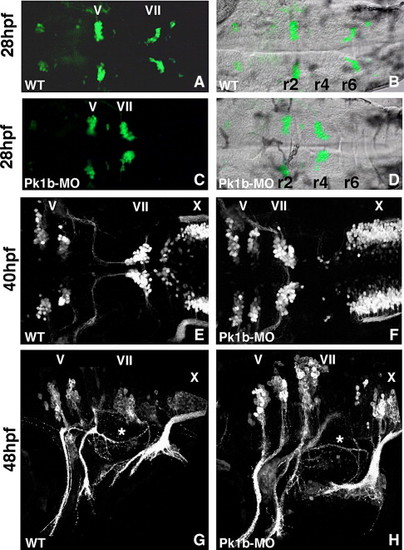
Pk1b function is required for facial neuron migration. The disposition of the branchiomotor neurons is visualized using islet1-GFP transgenic embryos, (A, C) viewed by fluorescence microscopy, (B, D) same image merged with bright field (DIC optics), or by confocal microscopy (E–H). The hindbrain is seen in dorsal (A–F) or lateral view (G, H); anterior is to the left in all panels. Rhombomeres (r) 2, r4, and r6 are labeled. (A, B) In wild-type 28 hpf embryos, the facial neurons (VII) have migrated most of the way to r6/7. (C, D) In Pk1b-deficient embryos (injected with Pk1b-MO), the facial neurons do not migrate and are instead found in clusters in r4. (E) In wild-type 40 hpf embryos, facial neuron migration is complete. (F) In 40 hpf Pk1b-deficient embryos, the facial neurons remain in r4. (G) In wild-type 48 hpf embryos, the facial neurons lie in r6/7, and their axons project out of r4 to innervate the second pharyngeal arch. The asterisk marks the axons of the VIIIth nerve/octavolateralis efferent (OLe), which project across the otic vesicle. (H) In Pk1b-deficient embryos, the facial (VII) and OLe (asterisk) axons can be seen to project normally. However, the facial neurons (VII) lie in r4, in an unusually dorsal position. MO: morpholino; V: trigeminal neurons; VII: facial neurons; X: vagal neurons. |
|
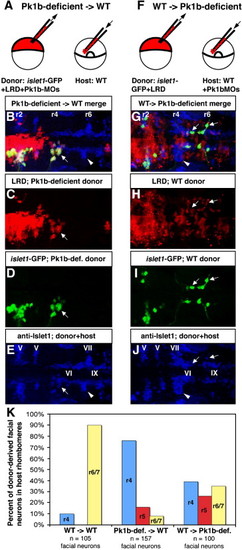
Transplantation reveals cell-autonomous function of Pk1b in facial neuron migration. (A) Schematic of cell transplantation approach to determine if Pk1b functions cell-autonomously in facial neuron migration. (B–E, G–J) Composite confocal Z-stacks of 36 hpf transplanted antibody-stained embryos in dorsal view; anterior is to the left. White arrows indicate donor-derived facial BMNs. White arrowheads indicate host-derived facial BMNs. Rhombomeres (r) 2, r4, and r6 are labeled. (B) Merged image (of C–E) of a transplanted embryo in which Pk1b-deficient (LRD-labeled, islet1-GFP-positive) donor cells have been transplanted into a wild-type host. (C) LRD labels all donor-derived cells. (D) Anti-GFP antibody specifically labels donor-derived islet1-GFP-positive BMNs. (E) Anti-Islet1 antibody (blue) shows both wild-type host facial BMNs in r6/7 (white arrowhead) and Pk1b-deficient donor-derived facial BMNs in r4 (white arrow; co-labeled with LRD and GFP, see B–D). Note that the anti-Islet1 antibody also labels the trigeminal (V) BMNs in r2 and r3, abducens (VI) neurons in r5 and r6, and glossopharyngeal (IX) BMNs in r7. (F) Schematic of cell transplantation approach to determine if Pk1b functions non-cell-autonomously in facial neuron migration. (G) Merged image (of H–J) of a transplanted embryo in which wild-type (LRD-labeled, islet1-GFP-positive) donor cells have been transplanted into a Pk1b-deficient host. (H) LRD labels all donor-derived cells. (I) Anti-GFP antibody specifically labels donor-derived islet1-GFP-positive BMNs. (J) Anti-Islet1 antibody (blue) shows both Pk1b-deficient host facial BMNs in r4 (white arrowhead) and wild-type donor-derived facial BMNs in r4–r7 (white arrows; co-labeled with LRD and GFP, see G–I). Note that the anti-Islet1 antibody also labels the trigeminal (V) BMNs in r2 and r3, abducens (VI) neurons in r5 and r6, and glossopharyngeal (IX) BMNs in r7. (K) Summary of transplant data showing percent of donor-derived facial BMNs located in each rhombomere in WT → WT, Pk1b-deficient → WT, and WT → Pk1b-deficient transplants. LRD: lysinated rhodamine dextran; MO: morpholino; V: trigeminal neurons; VI: abducens neurons; VII: facial neurons; IX: glossopharyngeal neurons. |
|
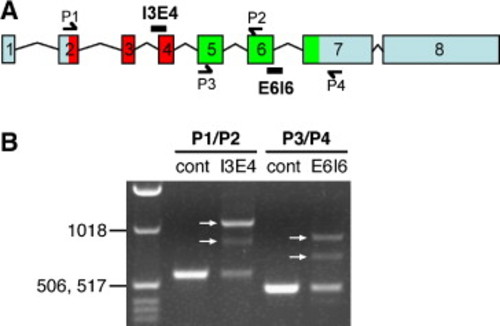
Pk1b splice-blocking morpholinos alter pk1b splicing. (A) Genomic organization of zebrafish pk1b; exons are numbered. Locations of the splice morpholinos Pk1b-MO-SA-I3E4 and Pk1b-MO-SD-E6I6, and locations of primers used to amplify the regions surrounding exon 4 (P1/P2) and exon 6 (P3/P4) are shown. As in Supplemental Fig. 1A, the region encoding the PET domain is colored red, and the region encoding the LIM domains is colored green. (B) RT–PCR analysis of pk1b mRNA in control uninjected and Pk1b-splice-MO-injected embryos. PCR primer combinations are shown above lanes. The ladder in the left lane includes bands of 1018 bp, 517 bp, and 506 bp. The region surrounding exon 4 was amplified from control embryos (cont) using P1 and P2, generating a PCR product of the expected size (582 bp). Amplification of the region surrounding exon 4 from Pk1b-MO-SA-I3E4-injected embryos (I3E4) generated two different splice variants (arrows, I3E4 lane). In one variant, the PCR product increased by the predicted size of intron 3 (515 bp). In the other variant, the PCR product increased in size, but by less than the predicted size of intron 3, suggesting the existence of a cryptic splice site within intron 3. The region surrounding exon 6 was amplified from control embryos (cont) using P3 and P4, generating a PCR product of the expected size (476 bp). Amplification of the region surrounding exon 6 from Pk1b-MO-SD-E6I6-injected embryos (E6I6) generated two different splice variants (arrows, E6I6 lane). In one variant, the PCR product increased by the predicted size of intron 6 (438 bp). In the other variant, the PCR product increased, but by less than the predicted size of intron 6, suggesting the existence of a cryptic splice site within intron 6. Overall, these data suggest that each splice-blocking morpholino blocks normal splicing, resulting in inappropriate inclusion of all or part of the adjacent intron. Aberrant translation of either intron 3 or intron 6 would result in a truncated protein due to a stop codon 5–6 amino acids into each intron. These results confirm the efficacy and specificity of both Pk1b-splice-blocking morpholinos (see Materials and methods for primer sequences). |
Reprinted from Developmental Biology, 309(2), Rohrschneider, M.R., Elsen, G.E., and Prince, V.E., Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b, 358-372, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.