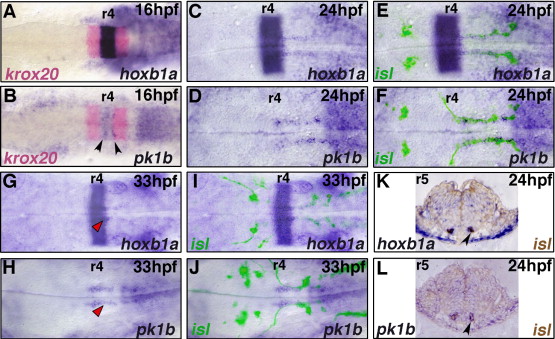
Fig. 5 hoxb1a and pk1b are expressed in migrating facial BMNs. In situ hybridization for hoxb1a (A, C, E, G, I, K) and pk1b (B, D, F, H, J, L) during facial BMN migration (blue); krox20 marks r3 and r5 (A, B, red). islet1-GFP (isl) expressing BMNs were labeled with anti-GFP antibody (fluorescence in panels E, F, I, J; brown staining in panels K, L). Embryos are flat-mounted with anterior to the left (A–J), or transverse 5 μm plastic sections through r5 (K, L). Rhombomeres (r) 4 and r5 are labeled. At 16 hpf, (A) hoxb1a and (B) pk1b are expressed in a stripe in r4; arrowheads in (B) indicate elevated pk1b expression in discrete domains within r4. By 24 hpf, (D) pk1b is no longer expressed in a stripe in r4, however both (C) hoxb1a and (D) pk1b are expressed in narrow bilateral trails stretching from r4 to r7. (E, F) This expression co-localizes with the facial BMNs along the length of their migration (C, D, bright-field; E, F same images merged with fluorescent anti-GFP label). (K, L) Co-localization is confirmed by transverse sections through r5. Arrowheads indicate facial BMNs labeled by both islet1-GFP (brown) and (K) hoxb1a, or (L) pk1b. At 33 hpf, (G) hoxb1a and (H) pk1b expression is significantly reduced in the facial BMNs, and is present in narrow bilateral domains in r4 and r5 which flank the facial neuron axon tracts marked by islet1-GFP (red arrowheads) (G, H, bright-field; I,J, same images merged with fluorescent anti-GFP label).
Reprinted from Developmental Biology, 309(2), Rohrschneider, M.R., Elsen, G.E., and Prince, V.E., Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b, 358-372, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.