Fig. 3
|
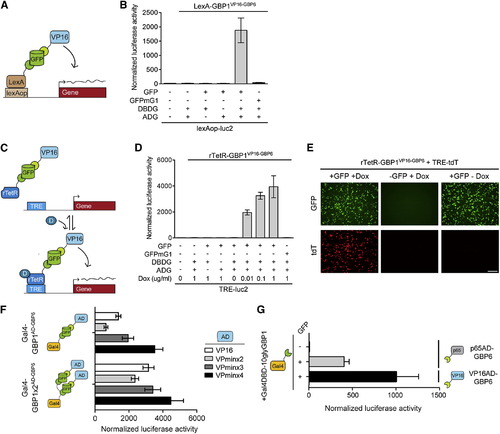
T-DDOGs Are Highly Adjustable (A–E) T-DDOGs based on LexA (A) and rTetR (C) DBDs. Doxycycline is “D” in (C). TRE includes seven tetO sequences (C). (B) LexA-GBP1VP16-GBP6 activated a lexAop-luc2 reporter only in the presence of GFP. n = 9. (D) rTetR-GBP1VP16-GBP6 activated TRE-luc2 in a GFP- and doxycycline-dependent manner. n = 6–9. (E) Similar results were seen with TRE-tdT. Doxycycline was used at 1 μg/ml. Images were taken 16 hr posttransfection. (F and G) Tuning T-DDOGs with adjustable DBDs and ADs. (F) Increasing the number of GBP1 on Gal4DBD (n = 6–9) enhanced the transcriptional potency for each ADG (n = 9). (G) Potency of p65AD compared to VP16AD. T-DDOGs used are Gal4-GBP1p65-GBP6 and Gal4-GBP1-BVP16-GBP6. n = 9. Scale bar, 100 μm. Plots are mean ± SD. |
Reprinted from Cell, 154(4), Tang, J.C., Szikra, T., Kozorovitskiy, Y., Teixiera, M., Sabatini, B.L., Roska, B., and Cepko, C.L., A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation, 928-939, Copyright (2013) with permission from Elsevier. Full text @ Cell