Fig. 2
|
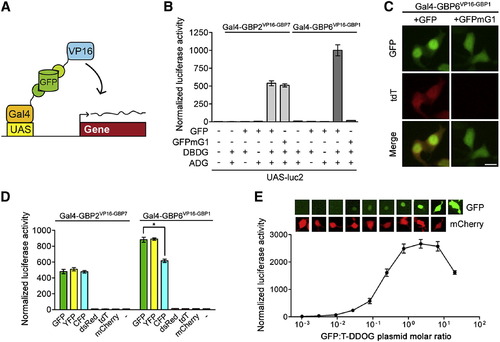
Characterization of the GFP-Dependent Transcription System (A) Schematic of Gal4-based T-DDOGs. (B) GFP-dependent activation of UAS-luc2 by Gal4-GBP6VP16-GBP1 and Gal4-GBP2VP16-GBP7. n = 9. (C) Gal4-GBP6VP16-GBP1 strongly activated UAS-tdT in the presence of GFP. Mutation of GBP1-binding residues in GFP (GFPmG1) abolished tdT activity. Scale bar, 10 μm. (D) Specificity of T-DDOGs for different fluorescent proteins. n = 9; p < 0.001. (E) Activity of Gal4-GBP6VP16-GBP1 in response to a varying amount of transfected GFP plasmids. The transfected DNA amount was kept constant among conditions, with CAG-mCherry (bottom) acting as a filler plasmid to compensate for reduction in GFP (top) plasmids. Panels show representative GFP and mCherry fluorescence in single cells for each corresponding data point below. n = 6. Plots are mean ± SD. See also Figures S1 and S2 and Table S2. |
Reprinted from Cell, 154(4), Tang, J.C., Szikra, T., Kozorovitskiy, Y., Teixiera, M., Sabatini, B.L., Roska, B., and Cepko, C.L., A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation, 928-939, Copyright (2013) with permission from Elsevier. Full text @ Cell