- Title
-
Distinct cellular and reproductive consequences of meiotic chromosome synapsis defects in syce2 and sycp1 mutant zebrafish
- Authors
- Olaya, I., Yilmaz, I.N., Nour-Kasally, N., Charboneau, R.E., Draper, B.W., Burgess, S.M.
- Source
- Full text @ PLoS Genet.
|
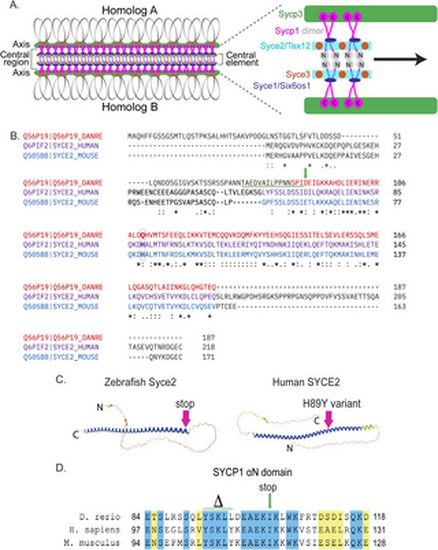
Generating the syce2 and sycp1 mutants. (A) Schematic of the synaptonemal complex showing key structures of meiotic chromosomes: DNA loops (gray/black), chromosome axis (Sycp3, green), central region (Sycp1, magenta) and the central element components (Syce2/Tex12, cyan; Syce1/Six6os1, dark blue; and Syce3, orange) that includes the Sycp1 N-termini (gray). (B) Alignment of the zebrafish Syce2 amino acid sequence with human and mouse orthologs using the Clustal Omega multiple sequence alignment tool [124]. Colored text defines regions that form a long α helix in a 2:2 complex [26]. Conserved identical residues (*), strongly similar (:) and weakly similar (.). The amino acids (aa) affected by the frameshift mutation are underlined in green. The position of the premature stop codon in syce2-/- is shown by a green arrow. The relative position of the human mutation (H89Y) associated with pregnancy loss [6] is highlighted in the boxed region. (C) AlphaFold-predicted structures for Syce2 protein from zebrafish (AF-Q56P19-F1) and human (AF-Q6PIF2-F1) [128]. The position of the 89th codon for zebrafish Syce2 that generates the premature stop codon is shown by a pink arrow. The position of the human variant in SYCE2 is depicted by the pink arrow. (D) Alignment of aa comprising the αN domain of Human SYCP1 that promotes self-assembly [25]. Blue regions are identical aa and yellow regions are similar aa. The CRISPR generated sycp1 mutation is a complex mutation with a deletion that removes the amino acids marked by delta and an insertion that truncates the protein at the amino acid marked by the arrow. See S1 Fig for details. |
|
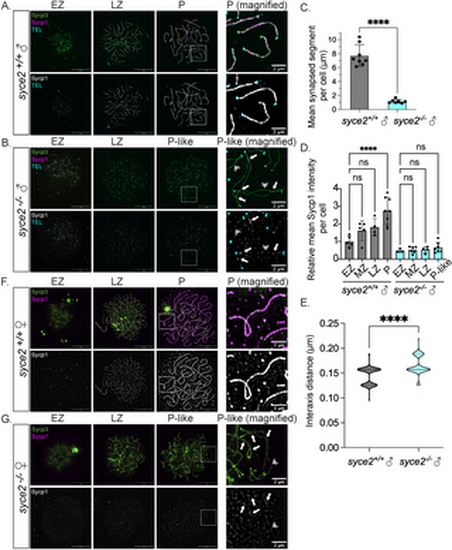
Loss of Syce2 function results in asynapsed interstitial regions. (A–B) Surface-spread chromosomes from syce2+/+ (A) and syce2-/- (B) spermatocytes imaged using structured illumination microscopy. Proteins are immunostained for Sycp3 (green) and Sycp1 (magenta and gray). Telomeres (cyan) are detected by in-situ hybridization using a fluorescent PNA probe. Examples of spread chromosomes during meiotic prophase I are shown: Early Zygotene (EZ), Late Zygotene (LZ), Pachytene (P) (syce2+/+), and Pachytene-like (P-like) (syce2-/-). Scale bar = 10 µm. Boxed region represents magnified examples of synapsed pachytene (syce2+/+) and pachytene-like (syce2-/-) chromosomes. Scale bar for magnified examples = 2 µm. Arrows represent synapsed-like regions with Sycp1 localization. Double arrowheads indicate Sycp1 localization at unaligned axial regions. (C) Mean synapsed segment per cell (µm) in pachytene and pachytene-like spermatocytes. The mean synapsed segment is significantly lower in syce2-/- spermatocytes (n = 7) compared to syce2+/+ (n = 8). Significance was determined using an unpaired t-test. **** = p < 0.0001. (D) Relative mean Sycp1 intensity per cell from early zygotene to pachytene or pachytene-like spermatocytes normalized to the mean syce2+/+ EZ values. Mean-fluorescence intensity between the axes (n = 25) – mean background (n = 6) was determined for each cell using the segmented-line tool in ImageJ. n = 5, syce2+/+ – EZ; n = 5; syce2+/+ – MZ; n = 4, syce2+/+ – LZ; n = 8, syce2+/+ – P; n = 3, syce2-/- – EZ; n = 6, syce2-/- – MZ; n = 4, syce2-/- – LZ; n = 7, syce2-/- – P-like. Significance was determined using ordinary one-way ANOVA testing with Šidák’s multiple comparisons test. ns = not significant; **** = p < 0.0001. (E) Distance between axes for co-aligned axial pairs in pachytene (n = 126) and pachytene-like (n = 93) spermatocytes from 8 cells in syce2+/+ and 7 cells in syce2-/- shown as a violin plot. Note that one pixel is equal to 0.03 µm resulting in a graph with a bimodal distribution. Significance was determined using an unpaired t-test. **** = p < 0.0001. (F–G) Surface-spread chromosomes from syce2+/+ (F) and syce2-/- (G) oocytes imaged using structured illumination microscopy immunostained for Sycp3 and Sycp1. Examples of spread chromosomes during meiotic prophase I are described in (A–B). Scale bar = 10 µm. Boxed region represents magnified examples of synapsed pachytene and pachytene-like chromosomes. Scale bar for magnified examples = 2 µm. Arrows represent synapsed-like regions with Sycp1 localization. Double arrowhead indicates Sycp1 localization at unaligned axial regions. |
|
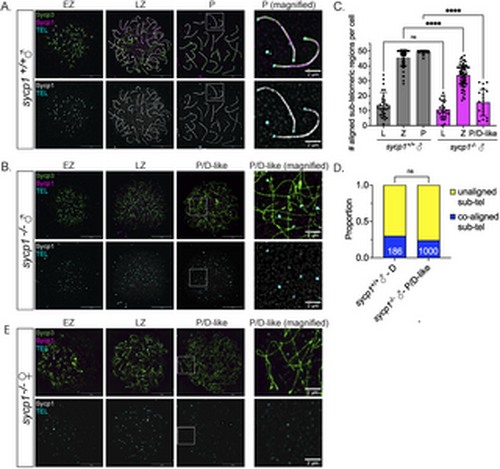
Formation and loss of co-alignment in sycp1-/- spermatocytes. (A) Surface-spread chromosomes from sycp1-/- spermatocytes imaged using structured illumination microscopy stained for Sycp3 (green), Sycp1 (magenta and gray) and telomeres (cyan). Examples of spread chromosomes during meiotic prophase I are shown: EZ (early zygotene), LZ (late zygotene), P (pachytene). Scale bar = 10 µm. Boxed region represents magnified examples of pachytene chromosomes. Scale bar for magnified examples = 2 µm. (B) Surface-spread chromosomes from sycp1-/- spermatocytes as described in (A); P/D (pachytene/diplotene-like). Scale bar = 10 µm. Boxed region represents magnified examples of P/D-like chromosomes. Scale bar for magnified examples = 2 µm. (C) Number of co-aligned pairs of sub-telomeric regions per cell across prophase I in sycp1+/+ and sycp1-/- spermatocytes from (A–B). Images for leptotene (L) are not shown but are characterized by predominantly short un-aligned axes. n = 27, sycp1+/+– L; n = 25, sycp1-/-– L; n, = 61 sycp1+/+– Z; n = 60, sycp1-/-– Z; n = 36, sycp1+/+– P; n = 20, sycp1-/-– P/D-like. Significance was determined using ordinary one-way ANOVA testing with Šidák’s multiple comparisons test. ns = not significant; **** = p < 0.0001. (D) Proportion of co-aligned and unaligned pairs of sub-telomeric regions in sycp1+/+ diplotene (D) (n = 186) and sycp1-/- P/D-like (n = 1000) spermatocytes. 11 cells were used for sycp1+/+ and 20 cells for sycp1-/-. Significance was determined using Fisher’s exact test. ns = not significant. (E) Surface-spread chromosomes from sycp1-/- oocytes as described in (A and B). Scale bar = 10 µm. Boxed region represents magnified examples of P/D-like chromosomes. Scale bar for magnified examples = 2 µm. |
|
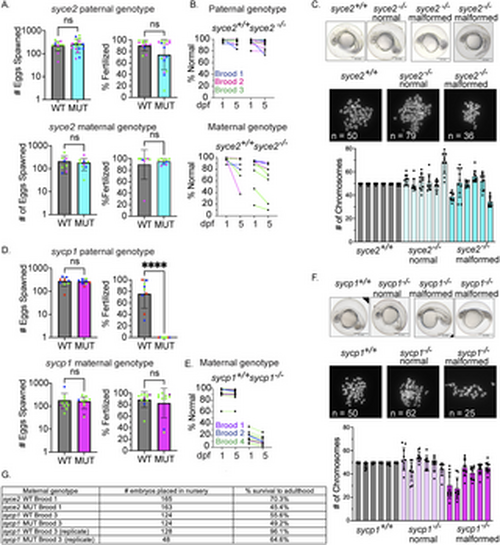
syce2-/- and sycp1-/- mutants have different reproductive outcomes. (A) Number of eggs spawned and fertilization rate per cross from wild-type and syce2-/- mutants; paternal genotype (top) and maternal genotype (bottom). Each data point represents a cross yielding > 20 eggs from individual animals from three different broods born on different days. Percent fertilized excludes decomposed eggs. Each dot represents an individual animal crossed to AB wild-type fish of the opposite sex. syce2 paternal genotype: Brood 1 (S4461; blue): WT n = 3, MUT n = 6; Brood 2 (S4562, pink): WT n = 2; MUT n = 5; Brood 3 (S4581; green): WT n = 6, MUT n = 6. syce2 maternal genotype: Brood 1 (S4461; blue): WT n = 2, MUT n = 1; Brood 2 (S4562, pink): WT n = 2; MUT n = 2; Brood 3 (S4581; green): WT n = 4, MUT n = 5. Significance was determined using an unpaired t-test for pooled data. ns = not significant. (B) Percent Normal-looking offspring at 1- and 5-days post fertilization (dpf) from wild-type and syce2 mutants from data in (A). Progeny from the syce2 paternal genotype (top) and progeny from the syce2 maternal genotype (bottom). Colored lines represent animals from 3 broods shown as colors in part A. (C) Examples of embryonic progeny of syce2+/+ mothers and normal-looking and malformed embryos from syce2-/- mothers (top). Examples of metaphase chromosome spreads from embryonic cells stained with DAPI isolated from embryos from syce2+/+ mothers and normal-looking and malformed embryos from syce2-/- mothers. The number of chromosomes in the spread is provided as n. Note, the embryos and spreads are independent and are not matched by animal. Quantification of the chromosome content in individual normal-looking (MUT-N) and malformed (MUT-D) offspring. Each column represents an individual embryo; dots represent the number of chromosomes in ten nuclei per embryo. Scale bar = 0.5 mm. (D) Number of eggs spawned and fertilization rate per cross for wild-type and sycp1-/- mutants; paternal genotype (top) and maternal genotype (bottom). Analysis was performed as described in (A). sycp1 paternal genotype includes Broods 2, 3, and 4. Brood 2 (S4297; blue): WT n = 4, MUT n = 3; Brood 3 (S4347; red): WT n = 3; MUT n = 2; Brood 4 (S44434; green): WT n = 2, MUT n = 4.The sycp1 maternal genotype includes Broods 1, 2, and 4. Brood 1 (S3667; purple): WT n = 2, MUT n = 1; Brood 2 (S4297; blue): WT n = 1; MUT n = 1; Brood 4 (S44434; green): WT n = 8, MUT n = 8. Significance was determined using an unpaired t-test for pooled data. ns = not significant.; **** = p < 0.0001. (E) Percent Normal-looking offspring at 1- and 5-days post fertilization (dpf) from wild-type and sycp1 maternal genotype. Colored lines represent animals from 3 broods in part D. (F) Analysis as in part C for progeny of sycp1+/+ and sycp1-/- mothers. (G) Table showing a subset of embryos placed in the nursery from select broods and the percent that survived to adulthood. |
|
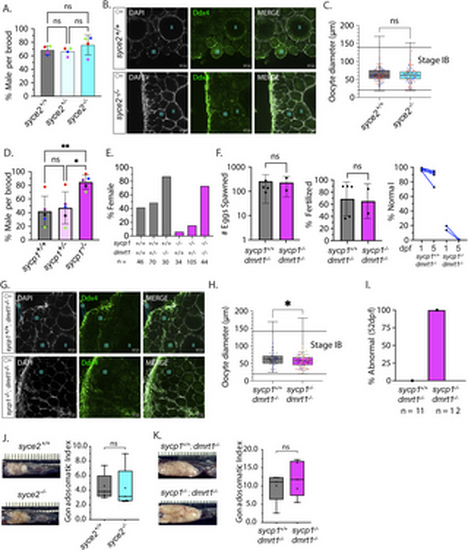
Normal sex ratios and gonad development in syce2-/- mutants but not sycp1-/- mutants. (A) Percentage of animals developing as male in syce2+/+, syce2+/- and syce2-/- genotypes from four independent experiments. Each data point indicates the percentage of males within each genotype out of total animals per brood: Blue (S4348) n= 99; Pink (S4461) = 342; Green (S4634) n = 318; Red (S4643) = 328. Significance was determined using repeated measures one-way ANOVA testing with Šidák’s multiple comparisons test; ns = not statistically significant. (B) Whole mount ovaries from 45 dpf syce2+/+ and syce2-/- females imaged using confocal microscopy. Ovaries are stained for DAPI (gray) and Ddx4 (green). Scale bar = 50 µm. (C) Calculated diameters of individual Stage IB oocytes from 45 dpf ovaries syce2+/+ (n = 3; 118 total oocytes) and syce2-/- (n = 3; 104 total oocytes). Different colors represent cells from different ovaries. Significance was determined using an unpaired t-test. ns = not statistically significant, * = p < 0.05. (D) Percentage of animals developing as male in sycp1+/+, sycp1+/- and sycp1-/- genotypes from five independent experiments. Each data point indicates the percentage of males within each genotype out of total animals per brood: Purple (S3667) n = 183; Blue (S4297) n= 237; Red (S4347) = 112; Black (S4395) n = 167; Green (S4434) n= 382. Significance was determined using repeated measures one-way ANOVA testing with Šidák’s multiple comparisons test; ns = not statistically significant, * = p < 0.05. (E) Percent females among genotypes of progeny from a sycp1-/+; dmrt1+/- incross. n values are given for each genotype. (F) Number of eggs spawned by sycp1+/+; dmrt1-/- (n = 5) and sycp1-/-; dmrt1-/- (n = 2) females (left) and the percent fertilized by AB males (center). Significance was determined using an unpaired t-test. ns = not statistically significant, * = p < 0.05. Percent of normal progeny (right) at 1 and 5 dpf for each cross. (G) Whole mount ovaries from 45 dpf sycp1+/+ and sycp1-/- females in a dmrt1-/- background stained with DAPI (gray) and Ddx4 (green). Scale bar = 50 µm. (H) Calculated diameters of individual Stage IB oocytes from two 45 dpf sycp1+/+; dmrt1-/- ovaries (n = 2; 81 total oocytes) and six 45 dpf sycp1-/-; dmrt1-/- ovaries (n = 6; 151 total oocytes). Different colors represent cells from different ovaries. Unpaired t-test; Significance was determined using an unpaired t-test. **** = p < 0.0001. (I) Percent abnormal ovaries by size by visual inspection dissected from 52 dpf fish. sycp1+/+; dmrt1-/- (n = 11) and sycp1-/-; dmrt1-/- (n = 12). (J) Ovaries from adult syce2+/+ and syce2-/- adult females (age = 11 months) in situ (left) and the gonadosomatic index of each (100 X gonad mass/total body mass) (right). N = 5 for each genotype. Unpaired t-test; ns = not statistically significant, * = p < 0.05. (K) Ovaries from sycp1+/+; dmrt1-/- and sycp1-/-; dmrt1-/- adult females (age = 13 months) as described in part J. N = 5 for each genotype. Unpaired t-test; ns = not statistically significant, * = p < 0.05. Note that the difference in ovary size from J is due to lower tank density and a greater amount of food per fish. |
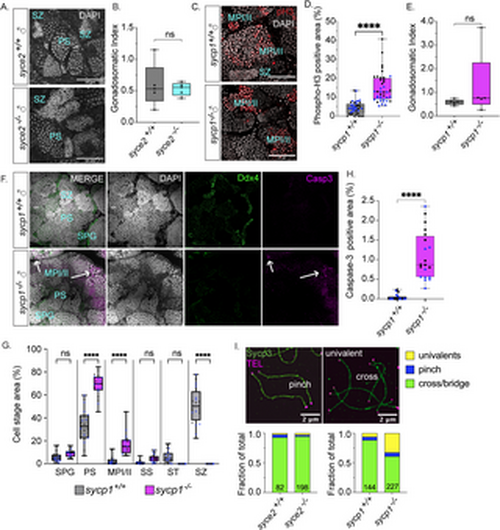
|
Spermatogenesis arrests at metaphase I/II in sycp1-/- males but not syce2-/- males. (A) Whole mount testes from adult (> 60 dpf) syce2+/+ and syce2-/- males stained with DAPI (gray). Examples of primary spermatocytes (PS) and spermatozoa (SZ). Scale bar = 50 µm. (B) The gonadosomatic index of syce2+/+ (n = 5) and syce2-/- (n = 5) testes. Unpaired t-test; ns = not statistically significant, * = p < 0.05. (C) Whole mount testes from adult (> 60 dpf) sycp1+/+ and sycp1-/- males stained with DAPI (gray) and phospho-Histone H3 (pH3; red). Examples of spermatocytes in metaphase I or II (MPI/II) and spermatozoa are noted. Scale bar = 50 µm. (D) Percent of cyst area represented by phospho-histone H3 staining. Multiple image fields (n = 20) were analyzed each for wild-type sycp1+/+ testes (n = 2) and sycp1-/- mutant testes (n = 2). Different colors represent data from same testis. Unpaired t-test; **** = p < 0.0001. (E) The gonadosomatic index of sycp1+/+ (n = 5) and sycp1-/- (n = 5) testes. Unpaired t-test; ns = not statistically significant, * = p < 0.05. (F) Whole mount testes from adult (> 60 dpf) sycp1+/+ (top) and sycp1-/- (bottom) males stained with DAPI (gray), Ddx4 (green) and cleaved caspase-3 (magenta). Arrows represent apoptotic nuclei. Scale bar = 50 µm. (G) Percent of cyst areas including spermatogonial cells (SPG), primary spermatocytes (PS), metaphase I/II (MPI/II), secondary spermatocytes (SS), spermatids (ST), and spermatozoa (SZ). Multiple image fields (n = 20) were analyzed each for wild-type sycp1+/+ testes (n = 2) and sycp1-/- mutant testes (n = 2). Measurements from individual testes shown in gray or blue circles. One-way ANOVA testing with Šidák’s multiple comparisons; **** = p < 0.0001; ns = not statistically significant, * = p < 0.05. (H) Percent of cyst area with cleaved caspase-3 positive cells. Multiple image fields (n = 20) were analyzed each for wild-type sycp1+/+ testes (n = 2) and sycp1-/- mutant testes (n = 2). Measurements from individual testes shown in black or blue circles. Unpaired t-test; **** = p < 0.0001. (I) Proportion of distinguishable chromosome configurations (cross/bridge bivalents, pinch bivalents and univalents) among diplotene (wild-type), pachytene-like (syce2-/-) and pachytene/diplotene-like (sycp1-/-) chromosomes. syce2+/+ (n = 82) and syce2-/- (n = 198); Chi-squared test: ns = not significant, ns = p < 0.05. sycp1-/- (n = 144) and sycp1+/+ (n = 227); Chi-squared p < 0.0001. Data is compiled from chromosomes spreads performed over seven independent experiments. |
|
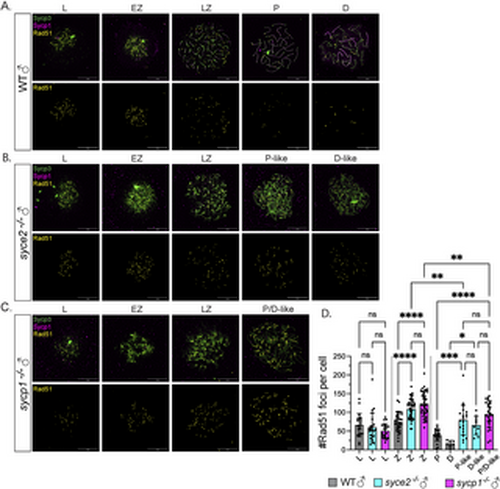
Inefficient DSB repair in syce2-/- and sycp1-/- mutants during prophase. (A–C) Surface-spread chromosomes from wild-type (A), syce2-/- (B) and sycp1-/- (C) spermatocytes immunostained for Sycp3 (green), Sycp1 (magenta) and Rad51 (yellow). Examples of spread chromosomes from meiotic prophase I: L, EZ, LZ, P, D, P-like (syce2-/-) and P/D-like(sycp1-/-). D-like nuclei (syce2-/-) are categorized as having full-length axes, but Sycp1 is largely absent from pseudo-synapsed regions throughout the nucleus. Scale bar = 10 µm. (D) Number of Rad51 foci per cell using data from (A–C). EZ to LZ nuclei are grouped together as zygotene (Z). Data is pooled from 2 independent experiments. n = 21, WT – L; n = 23, syce2-/- – L; n = 22, sycp1-/- – L; n = 34, WT – Z; n = 41, syce2-/- – Z; n = 44, sycp1-/- – Z; n = 22, WT – P; n = 7, WT – D; n = 21, syce2-/- – P-like; n = 10, syce2-/- – D-like; n = 26, sycp1-/- – P/D-like. Significance was determined using ordinary one-way ANOVA testing with Šidák’s multiple comparisons test. ns = not significant; ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001. |
|
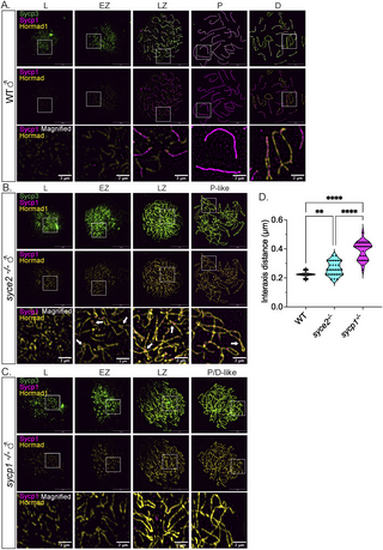
Sycp1 localization to synapsed-like regions is not sufficient for Hormad1 removal. (A–C) Surface-spread chromosomes from wild-type (A), syce2-/- (B) and sycp1-/- (C) spermatocytes immunostained for Sycp3 (green), Sycp1 (magenta) and Hormad1 (yellow). Examples of spread chromosomes from meiotic prophase I described in Fig 7A–C. Scale bar = 10 µm. Boxes represent magnified areas for each stage. Scale bar for magnified examples = 2 µm. Arrows represent Hormad1 localization to pseudo-synapsed regions with Sycp1. (D) Violin plot showing the distance between axes for co-aligned axial pairs in early zygotene for wild-type (n = 28), syce2-/- (n = 28) and sycp1-/- (n = 28) spermatocytes. 4 cells from each genotype were used for the analysis. Significance was determined using ordinary one-way ANOVA testing with uncorrected Fisher’s LSD test. ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001. |