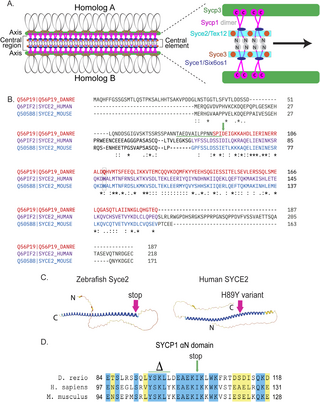
Fig. 1 Generating the syce2 and sycp1 mutants. (A) Schematic of the synaptonemal complex showing key structures of meiotic chromosomes: DNA loops (gray/black), chromosome axis (Sycp3, green), central region (Sycp1, magenta) and the central element components (Syce2/Tex12, cyan; Syce1/Six6os1, dark blue; and Syce3, orange) that includes the Sycp1 N-termini (gray). (B) Alignment of the zebrafish Syce2 amino acid sequence with human and mouse orthologs using the Clustal Omega multiple sequence alignment tool [124]. Colored text defines regions that form a long α helix in a 2:2 complex [26]. Conserved identical residues (*), strongly similar (:) and weakly similar (.). The amino acids (aa) affected by the frameshift mutation are underlined in green. The position of the premature stop codon in syce2-/- is shown by a green arrow. The relative position of the human mutation (H89Y) associated with pregnancy loss [6] is highlighted in the boxed region. (C) AlphaFold-predicted structures for Syce2 protein from zebrafish (AF-Q56P19-F1) and human (AF-Q6PIF2-F1) [128]. The position of the 89th codon for zebrafish Syce2 that generates the premature stop codon is shown by a pink arrow. The position of the human variant in SYCE2 is depicted by the pink arrow. (D) Alignment of aa comprising the αN domain of Human SYCP1 that promotes self-assembly [25]. Blue regions are identical aa and yellow regions are similar aa. The CRISPR generated sycp1 mutation is a complex mutation with a deletion that removes the amino acids marked by delta and an insertion that truncates the protein at the amino acid marked by the arrow. See S1 Fig for details.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ PLoS Genet.