- Title
-
Unique activities of two overlapping PAX6 retinal enhancers
- Authors
- Uttley, K., Papanastasiou, A.S., Lahne, M., Brisbane, J.M., MacDonald, R.B., Bickmore, W.A., Bhatia, S.
- Source
- Full text @ Life Sci Alliance
|
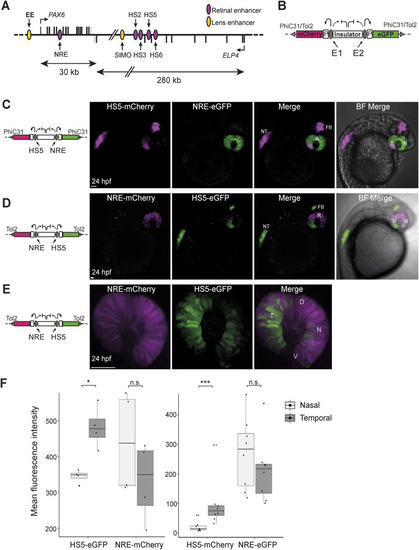
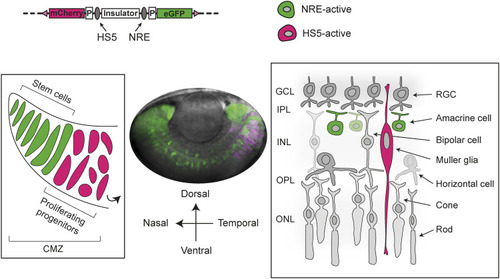
Activity of HS5 and NRE in a dual enhancer–reporter system during zebrafish embryonic development. |
|
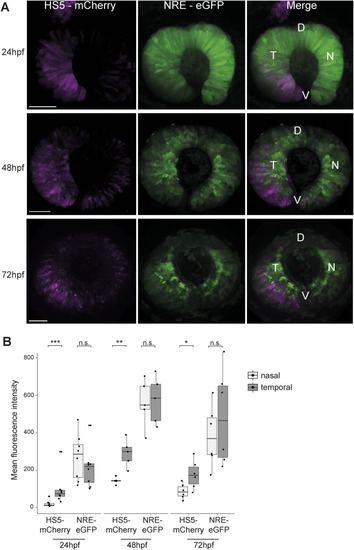
HS5 and NRE are active in different zones of the developing retina. |
|
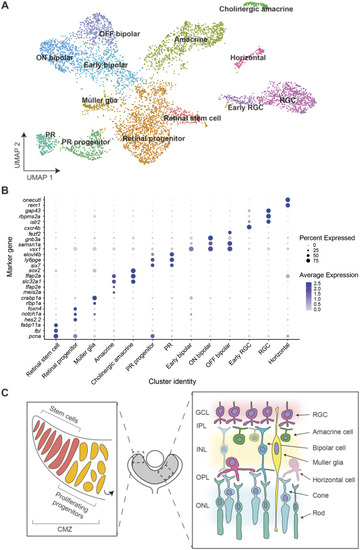
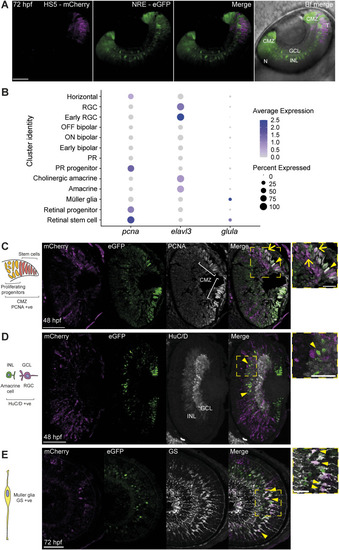
Single-cell RNA sequencing of NRE-eGFP/HS5-mCherry retinal cells at 48 hpf. |
|
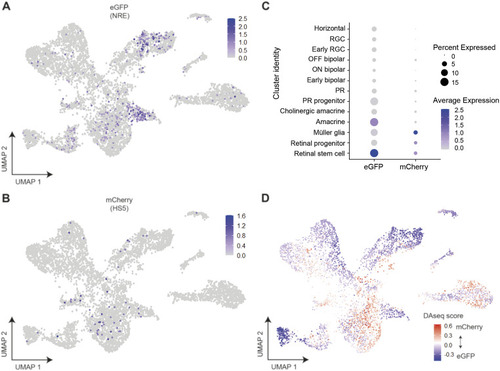
Assigning the identity of HS5 and NRE-active cells using the expression of eGFP and mCherry, and differential abundance analysis in cell-type clusters. |
|
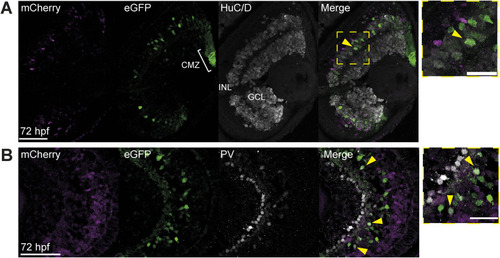
Immunofluorescence identifies enhancer-active cell types. |
|
|
|
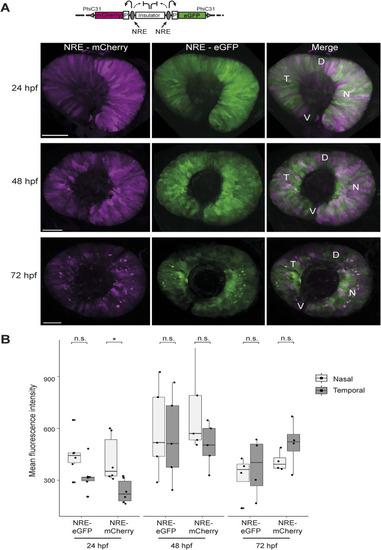
NRE control enhancer–reporter line reveals differential activity of NRE at 24 hpf. |
|
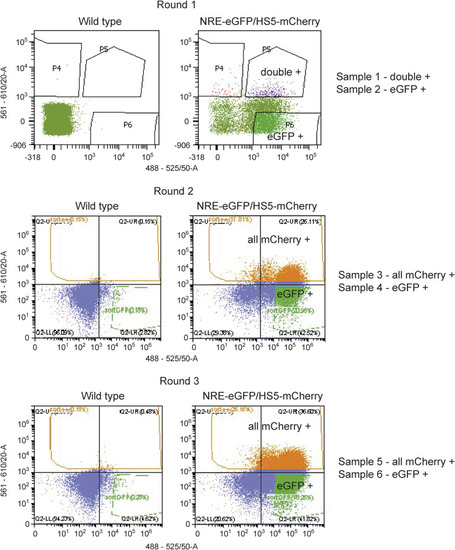
FACS profiles for scRNA-sequencing dataset. FACS plots showing WT controls (left) and NRE-eGFP/HS5-mCherry samples (right). Samples were sorted for eGFP (x-axis) and mCherry fluorescence (y-axis). In round 1, an eGFP-+ve sample and a double-+ve sample were collected and processed for scRNA-seq. In rounds 2 and 3, an eGFP-+ve sample and a sample of all mCherry-+ve were collected and processed for scRNA-seq. |
|
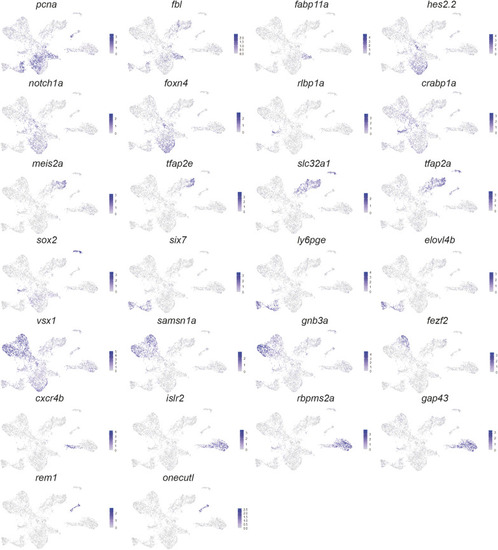
Expression of marker genes used for cluster annotation. Expression of cluster marker genes from |
|
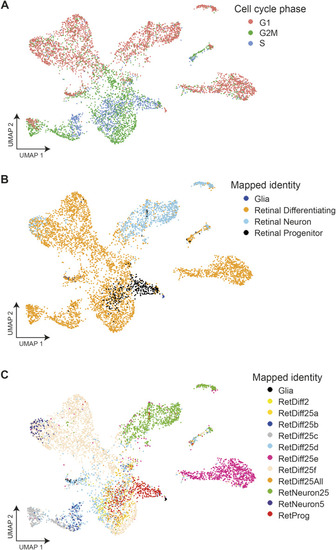
Mapped cell-type identities and assigned cell cycle phase. |
|
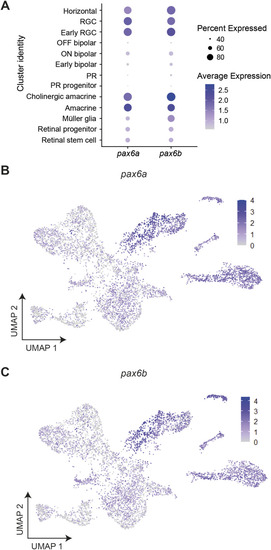
Expression of |
|
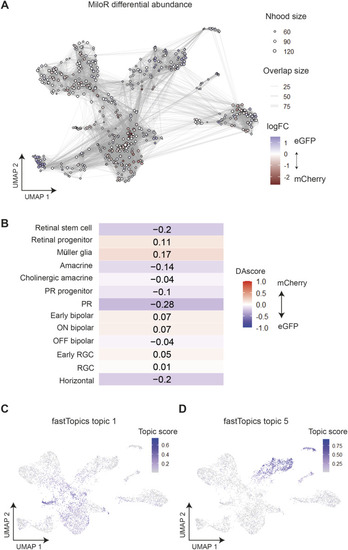
Differential abundance analysis and topic modelling validates the assignment of enhancer-active cell types. |
|
Immunofluorescence shows NRE-active amacrine cells. |