- Title
-
In vivo Reporter Assays Uncover Changes in Enhancer Activity Caused by Type 2 Diabetes Associated SNPs
- Authors
- Eufrásio, A., Perrod, C., Ferreira, F.J., Duque, M., Galhardo, M., Bessa, J.
- Source
- Full text @ Diabetes
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
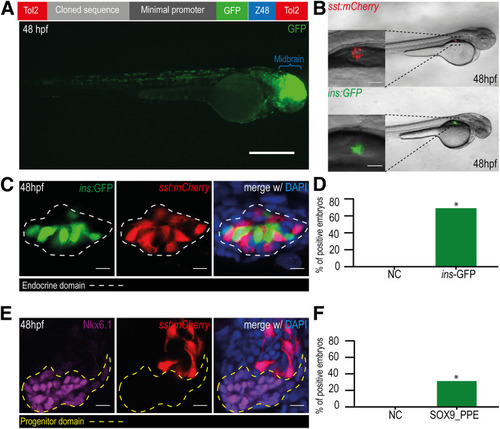
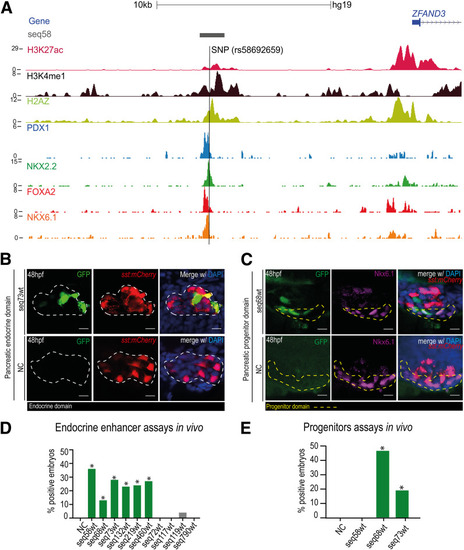
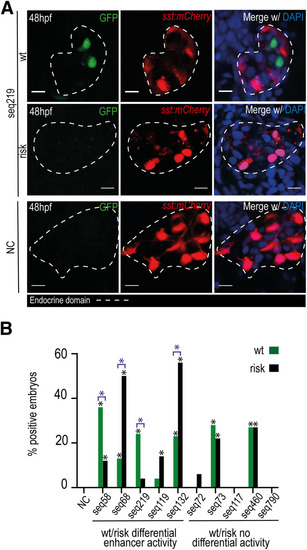
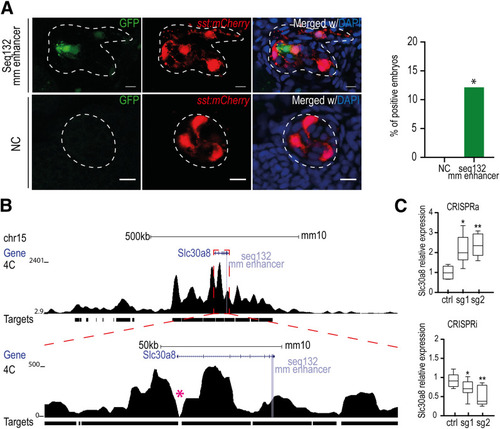
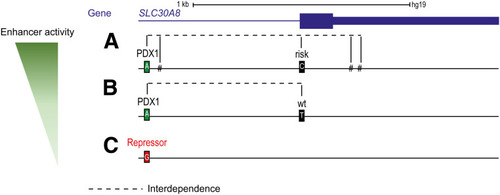
Theoretical model of the functional domains of seq132. Schematic representation from the three possible sequence versions and the respective associated enhancer activity level: high ( |