- Title
-
Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development
- Authors
- Schwend, T., and Ahlgren, S.C.
- Source
- Full text @ BMC Dev. Biol.
|
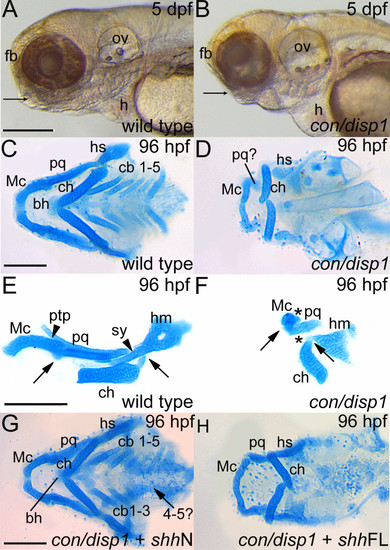
con/disp1 mutants display defective cartilage development in the PA which is partly rescued by RNA encoding non lipid-modified Shh. (A,B) Lateral views of live larvae show a reduction in head tissue and a deficit in jaw outgrowth (arrowhead) in con/disp1 mutant larvae at 5 dpf. (C) Ventral view of 96 hpf Alcian blue-stained wild type cartilages. (D) con/disp1 mutant cartilages reveal reduced, hypoplastic mandibular and hyoid arch cartilage elements and a complete absence of the cb 1-5 elements and midline-forming bh cartilage. (E,F) Lateral view of jaw cartilages reveals malformed joints (arrows denote joint site), fewer chondrocytes contributing to the pq cartilage element and a complete loss of the ptp and sy cartilage element in con/disp1 mutant (arrowhead denotes presence of cartilage, asterisk denotes cartilage absence). (G) Injection of 150 pg shhN mRNA into genotype-confirmed con/disp1 mutant mostly restores cartilage elements to wild type state (arrow indicates unilateral rescue in cb 4-5 elements), while (H) con/disp1 mutant injected with 150 pg shhFL mRNA are phenotypically similar to noninjected con/disp1 larvae. Scale bar: 50 μM. |
|
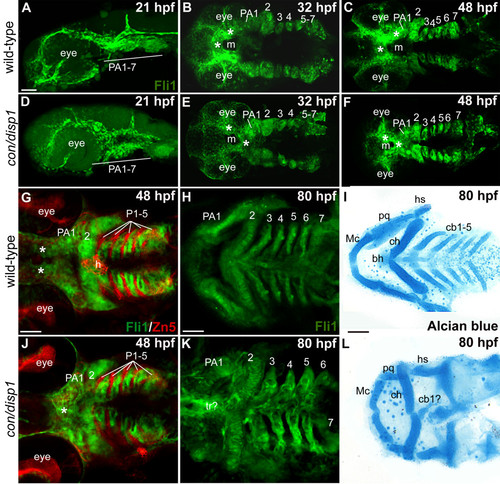
CNCC patterning in the con/disp1 mutant is visualized by the fli1gfp transgene. Lateral views (A,D) or ventral views (B,C,E-L) of confocal stack projections of fli1GFP+ CNCC in wild type and con/disp1 mutants (A-H, J,K) or Alcian blue-stained larvae to visualize cartilages (I,L). (G,J) Zn5 (red) staining to visualize endodermal pouches (P 1-5) in fli1GFP embryos. (A,D) At 21 hpf, wild type (A) and con/disp1 mutants (D) reveal similar patterns of postmigratory-CNCC. (B,C) 32 hpf ventral views of wild type embryos reveal CNC condensations in PA (B), and segmentation of CNCC by 48 hpf (C). (E,F) By 32 hpf, con/disp1 anterior-most CNCC (asterisks) become mispatterned, while CNCC within more PA 2-7 are correctly patterned (E) and segmented by 48 hpf (F). (G,J) CNCC in wild type (G) and con/disp1 mutants (J) are interdigitated properly in well-formed endodermal pouches 1-5 (P 1-5) (Zn5, red). (H,K) con/disp1 mutants show developed, segmented CNCC in posterior arches (K). (I,L) CNCC within con/disp1 mutant posterior arches fail to become cartilage. Scale bar: 50 μM. EXPRESSION / LABELING:
|
|
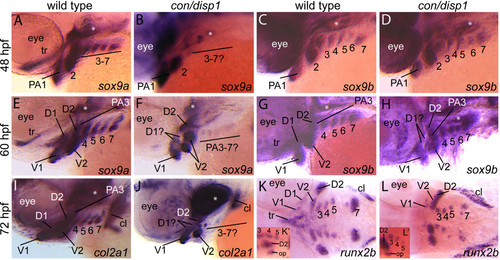
Chondrogenic differentiation in con/disp1 CNCC. Lateral views of 48 hpf (A-D), 60 hpf (E-H), 72 hpf (I,J) or ventral views of 72 hpf (K,L) wild type (A,C,E,G,I,K) or con/disp1 (B,D,F,H,J,L) embryos labeled with RNA probe for sox9a (A,B,E,F), sox9b (C,D,G,H), col2a1 (I,J) or runx2b (K,L). (A,C) sox9a marks CNCC in PA1-7 at 48 hpf in wild type embryos (A), however expression is lost in the posterior arch-residing CNCC in con/disp1 mutants (E). (C,D) sox9b continues to be expressed in 48 hpf PA1-7 condensations in wild type (B) and con/disp1 mutants alike (D). (E-H) At 60 hpf, sox9a and sox9b becomes downregulated in the dorsal subset of con/disp1 CNCC in the anterior arches (D1 and D2). sox9a remains absent in posterior arches in con/disp1 mutants, while sox9b remains robustly expressed in posterior arches. (I,J) At 72 hpf, col2a1 is reduced in con/disp1 D1 and D2 condensations and absent in PA3-7. (K,L) At 72 hpf, runx2b is absent in con/disp1 D1 and dorsal trabeculae precursors, but is present in PA condensation at wild type levels. (K′,L′) insets are of lateral views of same embryo shown in (K) and (L) respectively. Lateral views show presence of runx2b expression in opercle condensation in con/disp1 and wild type embryos alike. Asterisks indicate expression in mesoderm-derived polar cartilages. Images are at same magnification. EXPRESSION / LABELING:
|
|
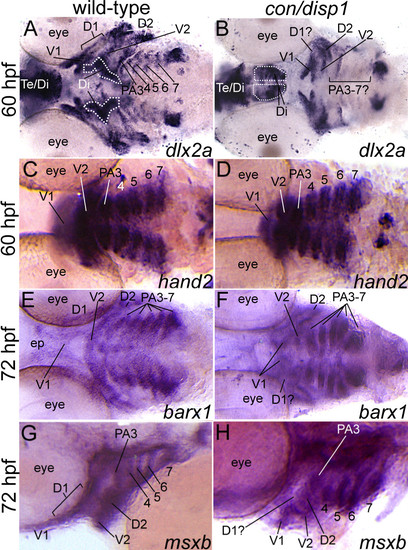
con/disp1 mutants show variable postmigratory-CNCC gene expression defects. Ventral, 60 hpf (A-D), ventral, 72 hpf (E,F) or lateral, 72 hpf (G,H) views of wild type (A,C,E,G) or con/disp1 (B,D,F,H) embryos labeled with RNA probe for dlx2a (A,B), hand2 (C,D), barx1 (E,F), msxb (G,H). (A,B) dlx2a is downregulated in dorsal domain of PA1 (D1) and posterior arches in con/disp1 mutants. (C,D) hand2 is expressed in CNCC within each con/disp1 PA. (E,F) At 72 hpf, barx1 expression marks the anterior and posterior arches in wild type embryos (E), but is downregulated in the con/disp1 D1 domain of PA1 (F). (G,H) msxb is downregulated in the D1 domain, but is expressed at wild type levels in posterior arch-residing CNCC. Scale bar: 50 μM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
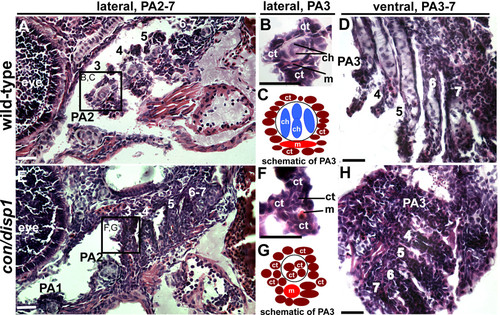
con/disp1 posterior arch-residing CNCC become fibrous-connective tissue. Lateral, (A,B,E,F) or ventral (D,H) H&E stained 10 μM sections of 5 dpf wild type (A,B,D) or con/disp1 mutant (E,F,H) larvae. (A,C) Chondrocytes are surrounded by connective tissue within each wild type PA (A), with higher magnification of PA3 revealing individual cell types (B). Box in (A) surrounding PA3 is viewed at higher magnification in (B). (E,F) con/disp1 mutants lack chondroctyes in PA3-7 (E), higher magnification of PA3 displays ectopic fibrous-connective tissue in PA3 (F). Box in (E) is viewed at higher magnification in (F). (C,G) Schematic of cell types visualized in wild type (C) and con/disp1 (G) PA3. (D,H) Horizontal sections show chondrocytes stacked within wild type posterior arches (D), while fibrous-connective tissue populate the con/disp1 posterior arches (H). Scale bar: 50 μM. PHENOTYPE:
|
|
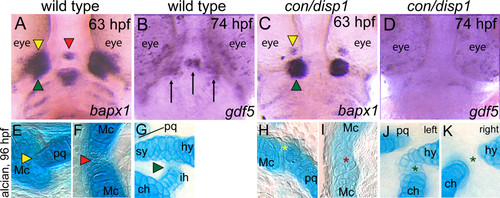
Joint defects are apparent in con/disp1 mutants. Ventral views (A-D) of wild-type (A,B) or con/disp1 mutants (C,D) labeled with RNA probe for bapx1 at 63 hpf (A,C) or gdf5 at 74 hpf (B,D). Alcian blue-stained cartilage at 96 hpf in wild-type (E-G) or con/disp1 mutant (H-K) embryos. (A,C) bapx1 prefigures bilateral and midline joints in wild-type embryos, but expression is reduced bilaterally and absent in the midline of con/disp1 mutants. Different color arrowheads used to denote individual cranial joints in A,C and E-G. Asterisks of the same color designate a reduced joint in con/disp1 mutants in H-K. (B,D) gdf5 is strongly expressed bilaterally in the first arch joint region (arrows) and in a group of cells in the midline that prefigures the basihyal cartilage element (arrow) (B). Expression of gdf5 is significantly reduced in both bilateral and midline domains in con/disp1 mutants (D). (E-G) In wild-type embryos, bilateral joints form between the Mc and pq (E, yellow arrowhead) and hs and ch (G, green arrowhead), as well as at the midline between the bilaterally formed Mc (F, red arrowhead). (H-K) In con/disp1 mutants, a poorly formed joint between the Mc, which fails to extend into a RAP element, and a severely reduced pq is visualized (H, yellow asterisk). In the second arch, we see either ectopic cartilage cells at the joint site between the hs and ch elements (J, green asterisk) or a failure of the hs and ch to join (K, green asterisk). Both phenotypes are occasionally present in the same larvae (left designates left side, right designates right side of single larvae). Further, the bilateral mc elements fuse at the midline to disrupt the joint (I, red asterisk). EXPRESSION / LABELING:
|
|
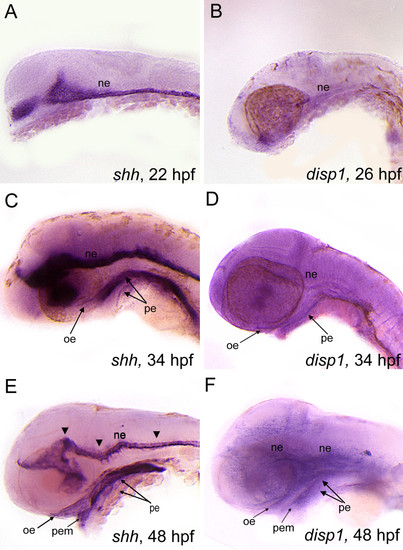
shh and disp1 are coexpressedin the developing head. Lateral views of wild type embryos labeled with RNA probe for shh (A,C,E) or disp1 (B,D,F). (A,B) At 22 hpf (shh) - 25 hpf (disp1)are both expressed in ventral neuroectoderm (ne). (C,D) At 34 hpf, shh and disp1 expression becomes detectable in oral ectoderm (oe) and pharyngeal endoderm (pe), in addition to the neuroectoderm (ne). Arrowheads denote expression within the neuroectoderm for both shh and disp1. (E-F) By 48 hpf, shh and disp1 expression becomes more prominent in the oe and pe, and is further expanded to the pharyngeal ectodermal margin (pem). shh and disp1 expression persists in the ne at 48 hpf. EXPRESSION / LABELING:
|
|
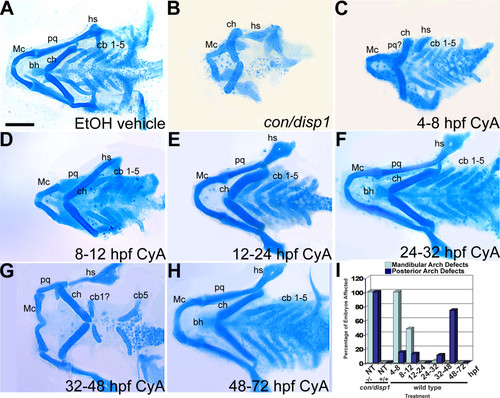
Early and late Cyclopamine treatments disrupt ventral PA development. (A-H) 96 hpf Alcian blue-stained cartilage. Wild type treated with ethanol (EtOH) vehicle (4-48 hpf) (A), untreated con/disp1 mutant (B) and wild type treated with 100 μM Cya at stages between 4-72 hpf (C-H). (B,C) 4-8 hpf treatment causes reduced jaw cartilage and outgrowth defects (C), similar to con/disp1 mutant (B), while hyoid and posterior arch cartilages are unaffected (C). (D-F) 8-32 hpf treatments had little effect on PA cartilage development, with occasional jaw cartilage reductions in 8-12 hpf treatment (D). (G) 32-48 hpf treatment eliminated most cb cartilages, without reducing anterior arch cartilage. (H) Treatments after 48 hpf have no effect on PA cartilage development. (I) Mandibular arch patterning defects and posterior arch chondrogenesis defects are summarized for each treatment (n = 20 embryos per treatment). Scale bar: 50 μM. |
|
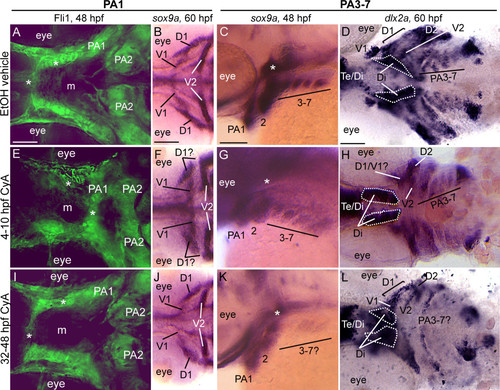
Early and late Cyclopamine treatments lead to CNCC defects. Ventral (A,B,D,E,F,H,I,J,L) or lateral (C,G,K) views at 48 hpf (A,C,E,G,I,K) or 60 hpf (B,D,F,H,J,L) of vehicle-treated wild type (A-D) or Cya-treated wild type (E-L) embryos that are Fli1gfp positive (A,E,I) or labeled with RNA probe for sox9a (B,C,F,G,J,K) or dlx2a (D,H,L). (A,E,I) By 48 hpf, vehicle-treated wild type embryos (A) display proper patterning of anteriormost CNCC (asterisks), as do 32-48 hpf Cya-treated embryos (I); however, 4-10 hpf Cya-treated embryos (E) show mispatterning of anteriormost CNCC. (B,F,J) Vehicle treated embryos display normal sox9a expression in the dorsal (D1) and ventral (V1) CNCC condensations in the first arch (B), as do 32-48 hpf Cya-treated embryos (J). In 4-10 hpf Cya-treated embryos, ventral (V1) CNCC condensations maintain sox9a expression, while sox9a is greatly reduced in the dorsal (D1) region of the first arch. (C,D,G,H,K,L) Treating wild type embryos with vehicle or Cya from 4-10 hpf does not influence sox9a expression at 48 hpf(C,G) or dlx2a at 60 hpf (D,H) within posterior arch residing CNCC, while treating wild type embryos with Cya at 32-48 hpf leads to reduction in sox9a gene expression at 48 hpf (K) and dlx2a at 60 hpf (L) within CNCC in the posterior arches. Asterisks in (C,G,K) indicate sox9a expression in mesoderm-derived polar cartilages. Scale bar: 50 μM. |
|
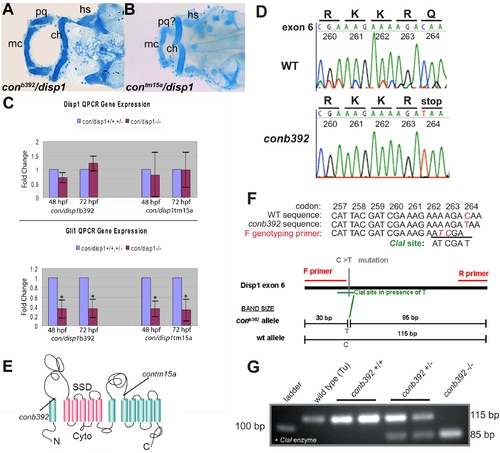
con/disp1b392 allele encodes a prematurely truncated dispatched1. (A,B) Ventral views of 4.5 dpf Alcian blue-stained cranial cartilage in con/disp1b392 (A) and con/disp1tm15a (B) larvae are phenotypically similar. (C) Both con/disp1 mutant alleles reveal a similar decrease in whole-embryo gene transcript levels of Hh-signaling responsive gene gli1 by QPCR. In contrast, disp1 RNA levels are similar between con/disp1 mutants and their wild type siblings, indicating that disp1 gene levels are not sensitive to Hh-signaling in whole embryos. (D) Sequence traces of disp1 from the con/disp1b392 allele reveal a premature stop codon in the sixth exon. (E) Schematic representation of the Disp1 protein. Arrows indicate sites of single base changes that introduce stop codons within disp1 gene as it corresponds to the mutant alleles used in this manuscript (con/disp1tm15a mutation described in [26]). (F,G) PCR genotyping strategy which utilizes a primer to introduce two unique nucleotides into Disp1 amino acids 262 and 263. These PCR generated mutations, when combined with the con/disp1b392 C to T transition in amino acid 264, generates a unique ClaI recognition site. Contrary to this, the ClaI site is not created in a wild type allele (F). The ClaI recognition site co-segregates with the con/disp1b392 mutant phenotype. Larvae were sorted at 48 hpf by characteristic mutant Shh-signaling axis defects, then genotyped for the ClaI RFLP by PCR and gel electrophoresis using primers within the genomic sequence of the sixth exon and subsequently digested by ClaI. ClaI digestion of the con/disp1b392mutant allele generates both 85 base pair (bp) and 30 bp fragments. ClaI fails to digest wild type alleles which results in an 115 bp fragment (G). The 30 bp fragments are not shown in (G) as they are hard to delineate within resulting primer dimers generated by the PCR and are unnecessary for our genotyping conclusions as the 85 bp fragment is sufficient. |
|
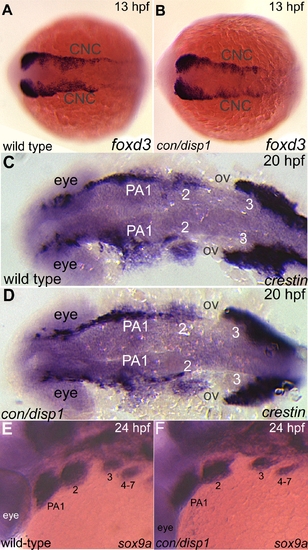
Specification and migration of CNCC occur normally in con/disp1 mutants. Dorsal (A-D), or lateral (E,F) views of 13 hpf (A,B), 20 hpf (C,D), or 24 hpf (E,F) wild type (A,C,E) or con/disp1 mutants (B,D,F) embryos labeled with RNA probe to foxd3 (A,B), crestin (C,D) or sox9a (E,F). (A-D) The patterns are indistinguishable between wild type and con/disp1 mutants at 13 hpf (foxd3) and 20 hpf (crestin). (E,F) sox9a marks CNC condensations at 24 hpf in wild type embryos (E) and con/disp1 mutants alike (F). |
|
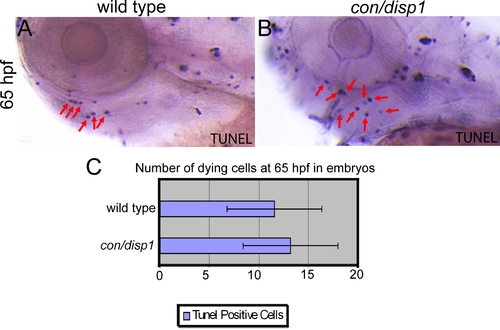
con/disp1 mutants do not display an increase in cell death within PA. (A,B) Lateral views displaying TUNEL positive cells in 65 hpf wild type (A) or con/disp1 (B) embryos. Red arrows in (A,B) are showing a portion of the cells we counted positive to reflect a dying cell in the pharyngeal region. The pharyngeal region was defined by us to include all tissue ventral to the eye and otic vesicle and dorsal to the heart and yolk. (C) Graph displaying the mean number of dying cells (11.6) for 11 wild type embryos (standard deviation was 4.8) and the mean number of dying cells (13.25) for 12 con/disp1 mutants (standard deviation was 4.8). This was not considered significant (p-value > 0.05, actual p-value was 0.432) by a two-tailed student t-test. These results reflect a lack of cell death at 65 hpf, which is consistent with what we saw at earlier and later stages (not shown in this figure). |
|
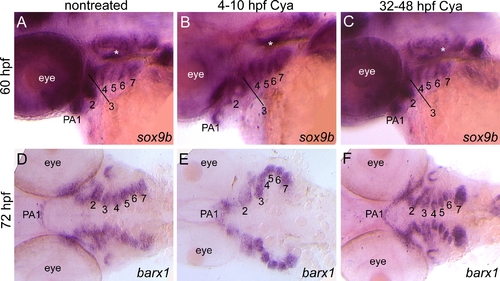
sox9b and barx1 expression following early and late Cya treatments. Lateral views of 60 hpf (A-C) or ventral views of 72 hpf (D-F) embryos that were untreated (A,D) or treated with Cya from 4-10 hpf (B,E) or 32-48 hpf (C,F) that are labeled with RNA probe for sox9b (A-C) or barx1 (D-F). (A-F) Cya treatments at these stages did not disrupt sox9b or barx1 expression patterns within mesenchymal condensation in PA. |