Fig. S1
- ID
- ZDB-FIG-091221-10
- Publication
- Schwend et al., 2009 - Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
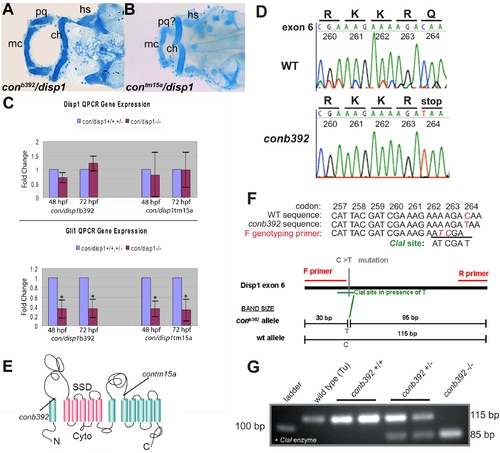
con/disp1b392 allele encodes a prematurely truncated dispatched1. (A,B) Ventral views of 4.5 dpf Alcian blue-stained cranial cartilage in con/disp1b392 (A) and con/disp1tm15a (B) larvae are phenotypically similar. (C) Both con/disp1 mutant alleles reveal a similar decrease in whole-embryo gene transcript levels of Hh-signaling responsive gene gli1 by QPCR. In contrast, disp1 RNA levels are similar between con/disp1 mutants and their wild type siblings, indicating that disp1 gene levels are not sensitive to Hh-signaling in whole embryos. (D) Sequence traces of disp1 from the con/disp1b392 allele reveal a premature stop codon in the sixth exon. (E) Schematic representation of the Disp1 protein. Arrows indicate sites of single base changes that introduce stop codons within disp1 gene as it corresponds to the mutant alleles used in this manuscript (con/disp1tm15a mutation described in [26]). (F,G) PCR genotyping strategy which utilizes a primer to introduce two unique nucleotides into Disp1 amino acids 262 and 263. These PCR generated mutations, when combined with the con/disp1b392 C to T transition in amino acid 264, generates a unique ClaI recognition site. Contrary to this, the ClaI site is not created in a wild type allele (F). The ClaI recognition site co-segregates with the con/disp1b392 mutant phenotype. Larvae were sorted at 48 hpf by characteristic mutant Shh-signaling axis defects, then genotyped for the ClaI RFLP by PCR and gel electrophoresis using primers within the genomic sequence of the sixth exon and subsequently digested by ClaI. ClaI digestion of the con/disp1b392mutant allele generates both 85 base pair (bp) and 30 bp fragments. ClaI fails to digest wild type alleles which results in an 115 bp fragment (G). The 30 bp fragments are not shown in (G) as they are hard to delineate within resulting primer dimers generated by the PCR and are unnecessary for our genotyping conclusions as the 85 bp fragment is sufficient. |