- Title
-
Deletion of the WD40 domain of LRRK2 in Zebrafish causes Parkinsonism-like loss of neurons and locomotive defect
- Authors
- Sheng, D., Qu, D., Kwok, K.H., Ng, S.S., Lim, A.Y., Aw, S.S., Lee, C.W., Sung, W.K., Tan, E.K., Lufkin, T., Jesuthasan, S., Sinnakaruppan, M., and Liu, J.
- Source
- Full text @ PLoS Genet.
|
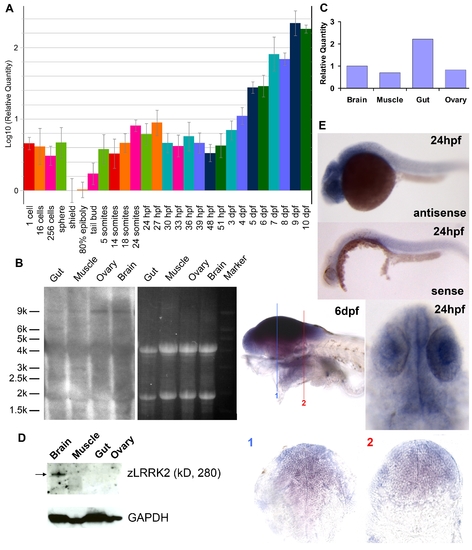
Expression profiling of lrrk2 in zebrafish. (A) Quantitative RT–PCR analysis of zlrrk2 mRNA expression from 1 cell stage to 10 dpf. (B–D) Northern blot analysis (B), quantitative RT–PCR (C), and Western blot analyses (D) of zLRRK2 expression in gut, muscle, ovary, and brain of adult fish. (E) In situ hybridization of zlrrk2 mRNA at 24 hpf and 6 dpf stage. For 6 dpf stage, cryo-sectioning was performed in the position labeled as 1 and 2. |
|
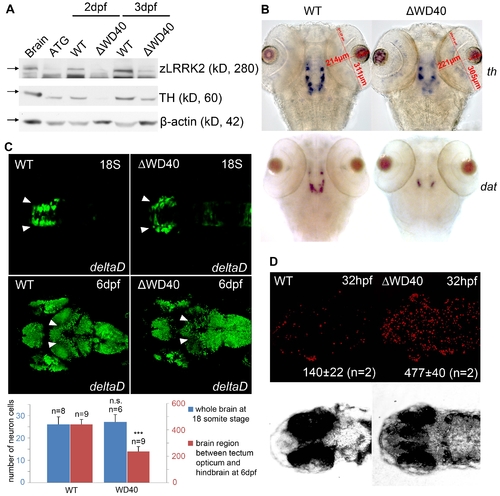
Phenotype of neuronal loss in LRRK2 ΔWD40 morphants. (A) Western blot analysis at 2 dpf and 3 dpf confirming the knockdown effect of zLRRK2 wild-type protein expression as well as the decreased TH protein expression in WD40 deletion morphant. Adult brain protein was used as positive control, and the ATG morphant at 3 dpf was used as negative control. The quantified results are shown in Figure S2. (B) WISH analysis at 3 dpf showing that WD40 domain deletion caused decreased TH and DAT expressions in WD40 deletion morphant, but without significant brain developmental retardation. (C) Uninjected (left) and WD40 deleted zebrafish expressing Kaede under the deltaD promoter. While neuron numbers is not much changed at 18 somite stage, there are fewer neurons in the midbrain of these 6 dpf fish, as indicated by the arrowheads. ***P<0.001, n.s.: not significant (unpaired Student′s t-test). (D) Cell apoptosis assay of uninjected (left) and WD40 delete zebrafish embryos. EXPRESSION / LABELING:
|
|
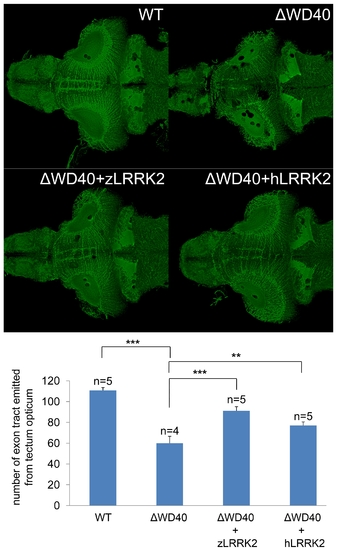
Analysis by acetylated tubulin staining of embryos at 6 dpf. Compared to the uninjected embryo, the WD40 deletion morphant has reduced and disorganized axon tracts in the midbrain of zebrafish. This phenotype could be rescued by over-expressing either zLRRK2 (WD40+zLRRK2) or hLRRK2 (WD40+hLRRK2) mRNA. **P<0.01, ***P<0.001 (unpaired Student′s t-test). |
|
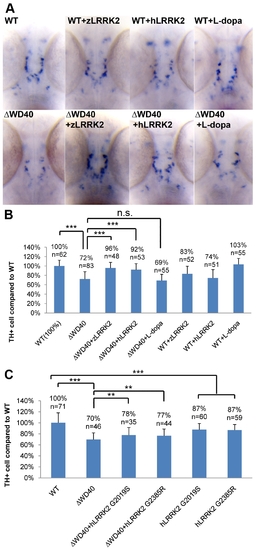
WISH Analysis of the loss of TH+ cell in WD40 morphant and its rescue by over-expressing wild-type zLRRK2 or hLRRK2 as well as hLRRK2 mutant alleles. (A) WISH analysis at 3 dpf shows the decreased TH expression in WD40 deletion morphant as well as the successful rescue by over-expressing either zLRRK2 or hLRRK2. The analysis also shows the decreased TH expression in the fish with over-expression of wild-type zLRRK2 and hLRRK2. This neurodegenerative phenotype could not be rescued by the treatment of L-dopa. (B) Quantification of rescue effect of zLRRK2 or hLRRK2 on TH+ cell loss in WD40 morphant. ***P<0.001 (unpaired Student′s t-test). (C) Quantification of rescue effect of hG2019S or hG2385R on TH+ cell loss in WD40 morphant. **P<0.01, ***P<0.001 (unpaired Student′s t-test). EXPRESSION / LABELING:
PHENOTYPE:
|
|
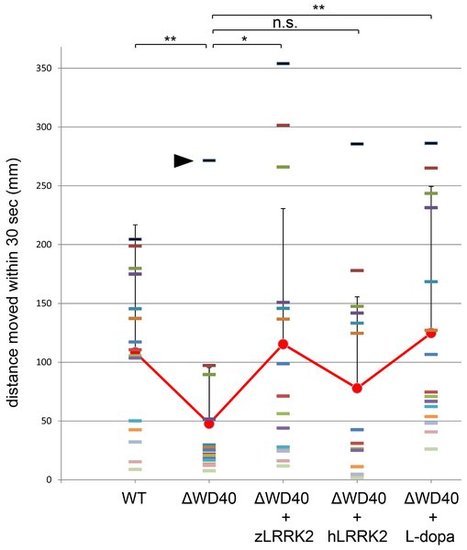
Analysis of locomotive movement by measuring the swimming distance in 30 seconds. The WD40 morphants swam much less distance than the wild-type fish. This locomotive defect could be rescued by over-expressing either zLRRK2 (WD40+zLRRK2) or hLRRK2 (WD40+hLRRK2) mRNA as well as by L-dopa treatment (WD40 + L-dopa). Each bar indicates the distance of an individual fish moved within 30 seconds. Each red dot (connected by red line) indicates the average distance of each of the five fish pools moved within 30 seconds, and the bar represents the SD of distance. *P<0.05, **P<0.01, n.s.: not significant (unpaired Student′s t-test). It is noticeable that one of the ΔWD40 fish showed a very different phenotype from the rest. The distinct phenotype of this ‘outlier’ fish was likely due to either the failure or in-efficient knock-down effect by morpholino. The rescue effect by over-expressing hLRRK2 became statistically significant (P = 0.03) when the possible outlier of the ΔWD40 group (black arrow) is removed from the statistical analysis. PHENOTYPE:
|
|
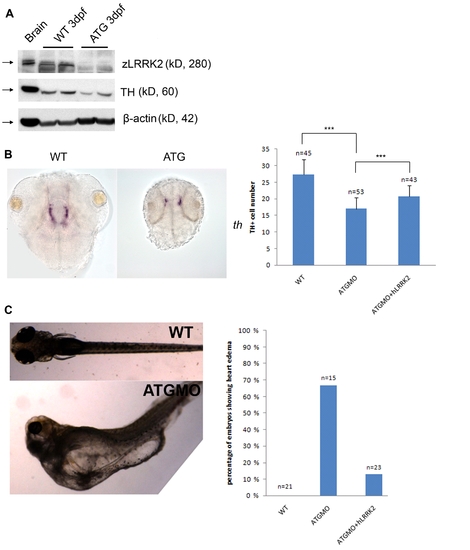
Phenotypes of zLRRK2 ATG morphants. (A) Western blot analysis at 72 hpf showing the strong reduction in expression of zLRRK2 protein as well as the decreased TH protein level in embryos injected with ATG morpholino. The analysis was done in duplicate, and adult brain protein was used as positive control for zLRRK2. (B) WISH analysis at 2 dpf showing retarded brain development and decreased TH expression in an ATG morphant (right). An uninjected sibling is shown for comparison (left). Quantification of the TH+ cell loss in ATG morphant as well as the rescue of ATG morphant with hLRRK2 was shown on the right. Co-injection of hLRRK2 significantly rescue the number of TH+ cells compared to ATG MO alone. ***P<0.001 (unpaired Student′s t-test). (C) Heart edema phenotype of ATG morphant and rescue effect of hLRRK2 on ATG morphant. |
|
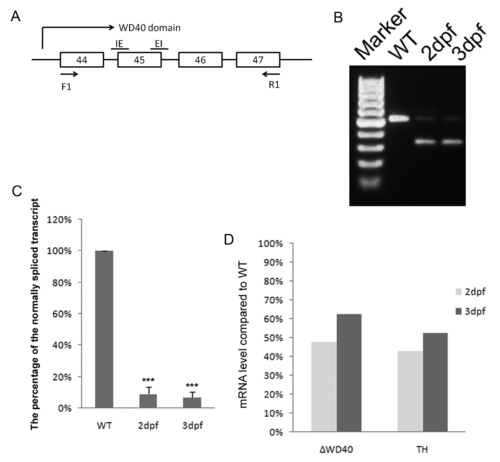
RT-PCR Analysis to confirm the blockage of exon 45 splicing by WD40 morpholino. (A) Schematic representation of the exons 44 to 47 of zLRRK2 to show the morpholinos used for blocking the 45th exon splicing (the track above the exons) as well as the primers (the track below the exons) used in the RT-PCR verification of the splicing blockage effect at 2 dpf and 3 dpf. (B) RT-PCR analysis at 2 and 3 dpf showing that the WD40 morpholino could block the splicing of exon 45, and the major of transcripts were the abnormally spliced one without exon 45 (confirmed by sequence analysis). (C) The percentage of the normally spliced transcript in the total amount of normal and abnormally spliced (without exon 45) transcripts in WT and WD40 morphant at 2 and 3 dpf. The densities of normal and abnormal spliced transcripts (in B) at 2 and 3 dpf were measured, and the percentage of normally spliced transcript was calculated by comparing the density of normal transcript to the total density of both normal and abnormal transcripts (sum of density from normal and abnormal forms). The percentage is presented as mean ± SD from 4 independent experiments. In wild-type embryos, no abnormal splicing forms were detected. ***P<0.001 (unpaired Student′s t-test). (D) Quantitative RT-PCR analysis of wild-type controls and WD40 morphants at 2 and 3 dpf, showing that both TH and WT zLRRK2 mRNA levels were reduced to 40%–60% of WT fishes. |
|
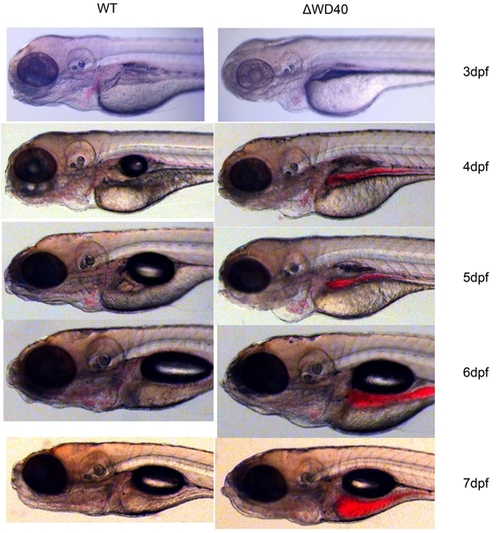
Morphological phenotype of WD40 morphants (From 3 dpf to 7 dpf). Morphants show no significant morphological defects compared to WT fishes except the mild blood accumulation in gut and pronephric duct between the york sac and swimming bladder. |
|
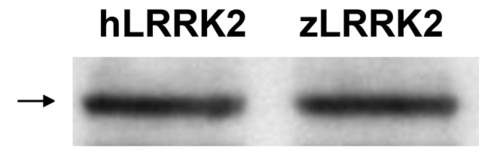
Linearized plasmid harboring either zLRRK2 or hLRRK2 cDNA tagged by Flag can be expression in zebrafish embryos by microinjection. |
|
Morphological phenotype of hG2019S and hG2385R overexpression. At 6 dpf, hG2019S and hG2385R overexpression shows no significant morphological defects compared to WT except the blood accumulation in gut and pronephric duct between york sac and swimming bladder. |
|
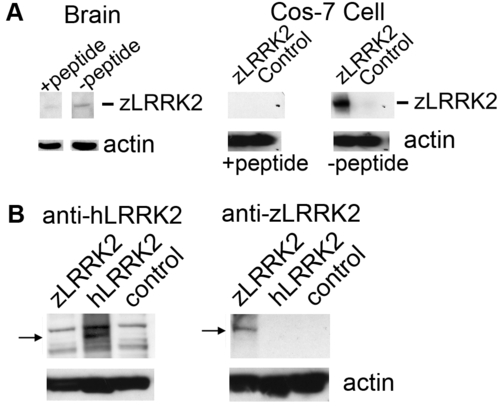
Analysis of zLRRK2 antibody specificity. (A) Specificity of zLRRK2 antibody specificity was verified by western blot analysis. Positive signals could be largely blocked by pre-incubating the anti-zLRRK2 antibody with the neutralizing peptide (+ peptide, left panel). Similarly, the zLRRK2 signal was completely blocked by pre-incubating the anti-zLRRK2 antibody with the neutralizing peptide in the zLRRK2-overexpressing Cos-7 cells (+ peptide, right panel). (B) Western blot analysis showing the specificity of the zLRRK2 antibody against zLRRK2 in Cos-7 cell line. Human LRRK2 and zLRRK2 recombinant proteins were overexpressed in Cos-7 cells separately and detected by anti-hLRRK2 (left panel) and anti-zLRRK2 (right panel), respectively. A positive band (280 kD, arrow) was only detected in hLRRK2-overexpressing cells (hLRRK2, left panel), using anti-hLRRK2 antibody (NOVUS NB 300–268). No obvious band was detected in zLRRK2-overexpressing and untransfected control cells (zLRRK2 and control, left panel); On the other hand, a sharp band (arrow) was detected in zLRRK2-overexpressing cells using the anti-zLRRK2 antibody (zLRRK2, right panel), but not in hLRRK2-overexpressing and untransfected control cells (hLRRK2 and control, right panel). This indicates the anti-zLRRK2 antibody is specifically against zLRRK2 proteins. |