|
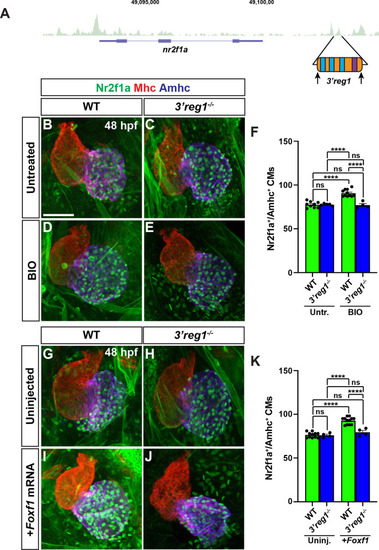
Wnt signaling and Foxf1 require 3’reg1 to promote a surplus ACs. A) Schematic of 3’reg1 enhancer deletion generated using CRISPR/Cas12a. Arrows indicate location of the guides. B-E) Confocal images of hearts from untreated and BIO-treated WT and 3’reg1Δ/Δ embryos stained for Nr2f1a (green), Mhc (red), and Amhc (blue). BIO was unable to promote an increase in ACs in 3’reg1-/- embryos. F) The number of Nr2f1a+/Amhc+ cardiomyocytes in untreated and BIO-treated WT and 3’reg1-/- embryos. Untreated WT (n = 9); untreated 3’reg1-/- (n = 5); treated WT (n = 11); treated 3’reg1-/- (n = 4). G-J) Confocal images of hearts from uninjected and Foxf1 mRNA-injected WT and 3’reg1-/- embryos stained for Nr2f1a (green), Mhc (red), and Amhc (blue). K) The number of Nr2f1a+/Amhc+ cardiomyocytes in uninjected and Foxf1 mRNA-injected WT and Nr2f1a+3’reg1-/- embryos. Uninjected WT (n = 11); Uninjected 3’reg1-/- (n = 4); Injected WT (n = 12); Injected 3’reg1-/- (n = 4). Scale bar: 50 μm. Error bars in graphs indicate s.e.m. **** indicate P < 0.0001. ns indicates not a statistically significant difference.
|