|
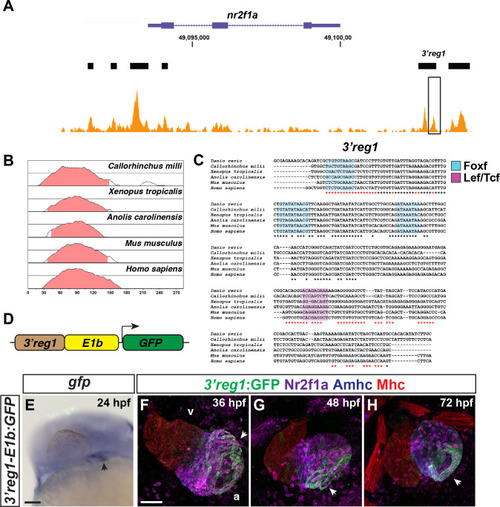
The 3’reg1-nr2f1a enhancer is expressed in the atrium. A) ATAC-seq profiling from ACs showing regions of open chromatin near the nr2f1a locus (purple). Black bars indicate called regions of open chromatin. Black box indicates 3’reg1. B) VISTA plot showing conservation of zebrafish 3’reg1 enhancer in Callorhincus milli (Australian ghostshark), Xenopus tropicalis (Tropical clawed frog), Anolis carolinensis (Green Anole), Mus musculus (House mouse), and Homo sapiens (human). Pink indicates >50% conservation of regulatory regions with zebrafish 3’reg1. Median lines in individual VISTA plots indicate 75% conservation. C) Clustal alignment of 3’reg1 from the VISTA plot in indicated species. Black asterisks indicate conserved nucleotides. Red asterisks indicate partially conserved nucleotides. Conserved putative binding sites for Foxf (blue shade) and Lef/Tcf (purple shade) TFs. D) Schematic of the 3’reg1:GFP reporter vector. E)In situ hybridization (ISH) for gfp in transgenic 3’reg1:GFP embryo at 24 hpf (n = 24). Venous pole of the heart tube (black arrowhead). View is lateral with anterior left and dorsal up. Scale bar: 200μm. F-H) Confocal images of hearts from transgenic 3’reg1:GFP embryos stained for 3’reg1:GFP (green), Nr2f1a (magenta), Amhc (ACs–blue), and Mhc (pan-cardiac–red). 36 hpf (n = 4), 48 hpf (n = 4), 72 hpf (n = 4). v–ventricle. a–atrium. 3’reg1:GFP in the atria of the hearts (white arrowheads). Images are frontal views with the arterial pole up. n indicates the number of embryos examined for representative experiment. Scale bar: 50 μm. **** indicate P < 0.0001.
|