- Title
-
Molecular blueprints for spinal circuit modules controlling locomotor speed in zebrafish
- Authors
- Pallucchi, I., Bertuzzi, M., Madrid, D., Fontanel, P., Higashijima, S.I., El Manira, A.
- Source
- Full text @ Nat. Neurosci.
|
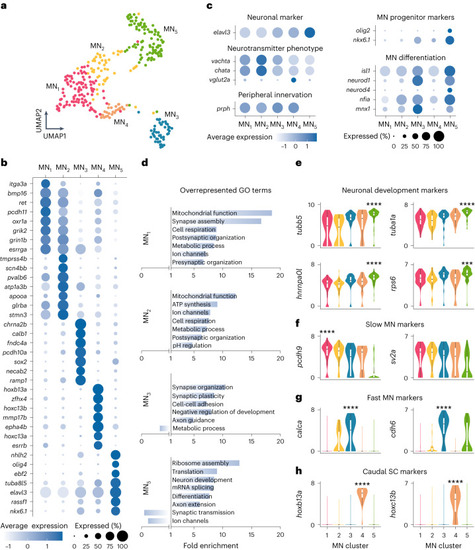
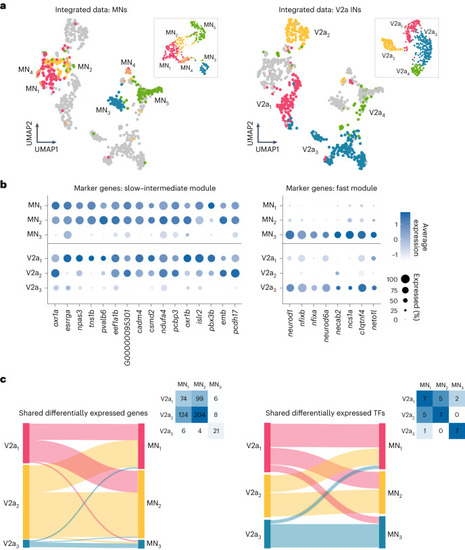
Molecular characterization of MN diversity. |
|
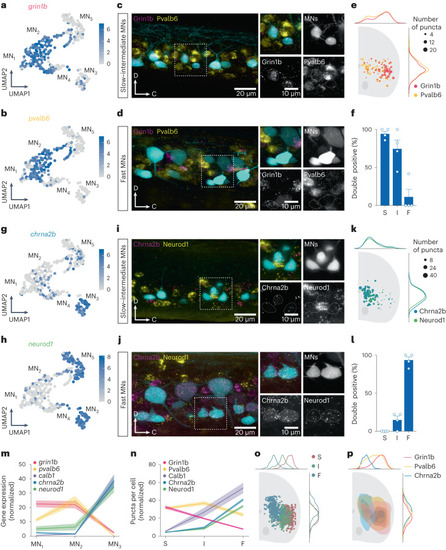
Validation of new molecular markers for MN subtypes. |
|
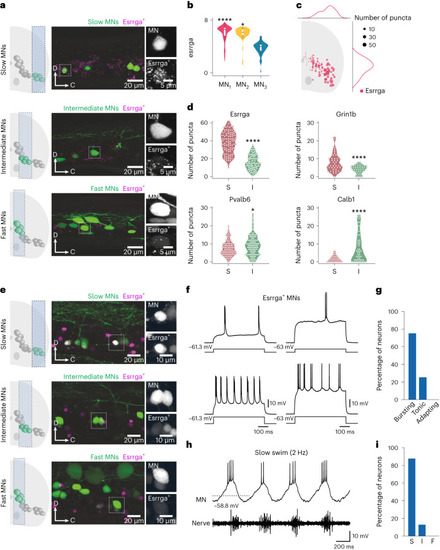
Characterization of Esrrga+ MNs. |
|
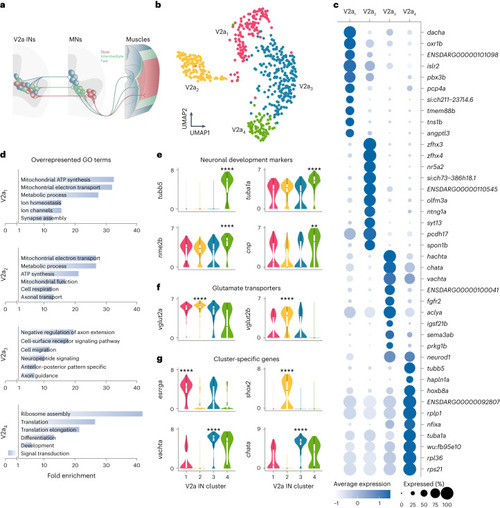
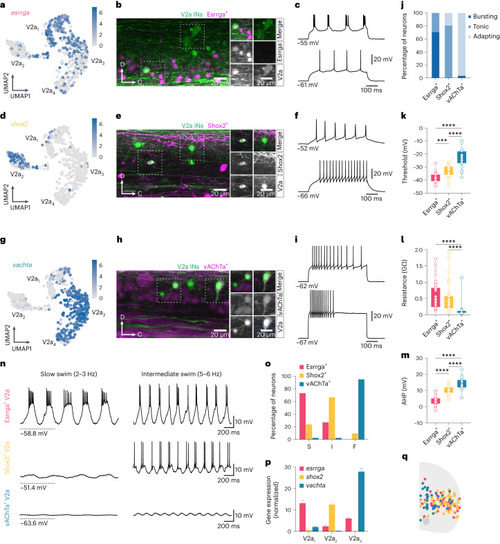
Molecular characterization of V2a IN diversity. |
|
Electrophysiological and functional validation of V2a IN molecular clusters. |
|
Shared features of functional speed modules across spinal populations |
|
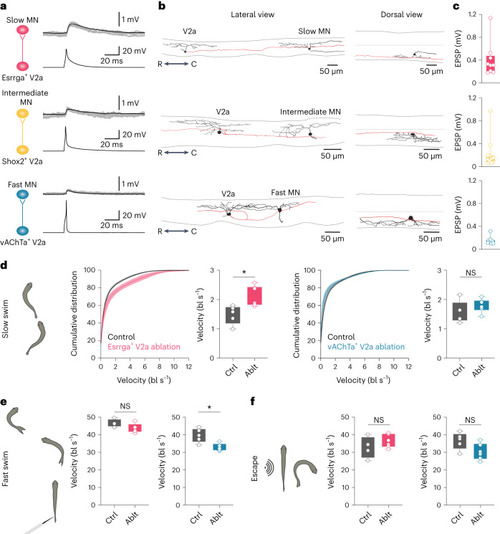
Functional connectivity between transcriptomically defined V2a IN and MN subtypes. |