- Title
-
Lineage tracing identifies heterogeneous hepatoblast contribution to cell lineages and postembryonic organ growth dynamics
- Authors
- Unterweger, I.A., Klepstad, J., Hannezo, E., Lundegaard, P.R., Trusina, A., Ober, E.A.
- Source
- Full text @ PLoS Biol.
|
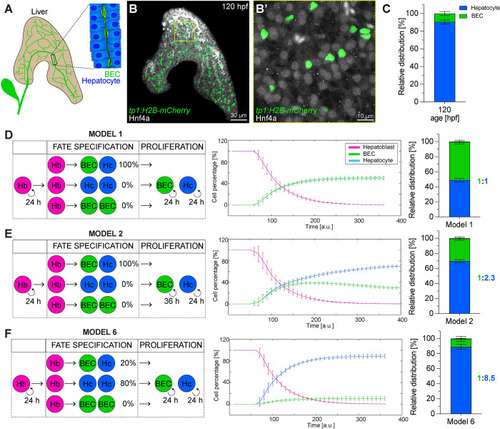
Establishment of BEC and hepatocyte lineages: in vivo cell type quantification and in silico modelling ( |
|
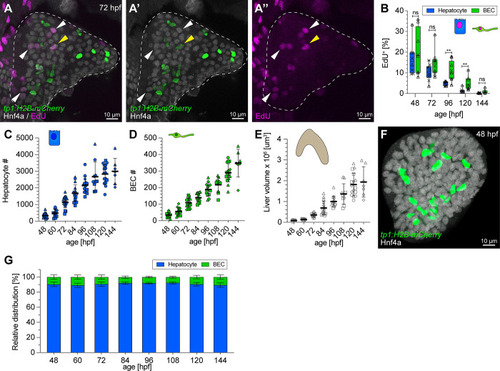
Hepatic proliferation dynamics and early establishment of a 1:9 BEC:hepatocyte ratio during embryonic development. ( |
|
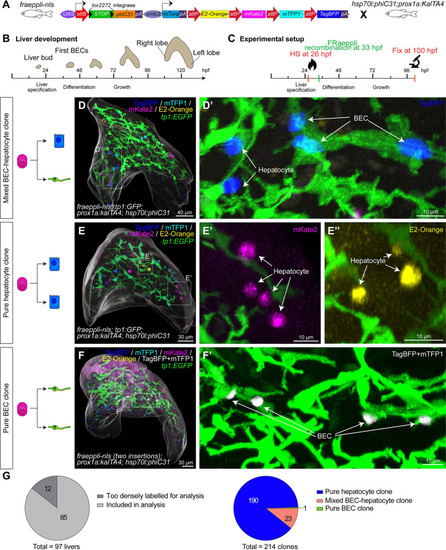
Quantitative lineage tracing identifies uni- and bipotent hepatoblast contributions during lineage decisions. ( |
|
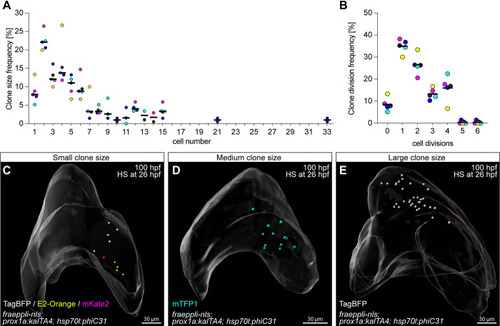
Quantitative lineage tracing of hepatoblasts during embryonic development identifies heterogeneous growth behaviour. ( |
|
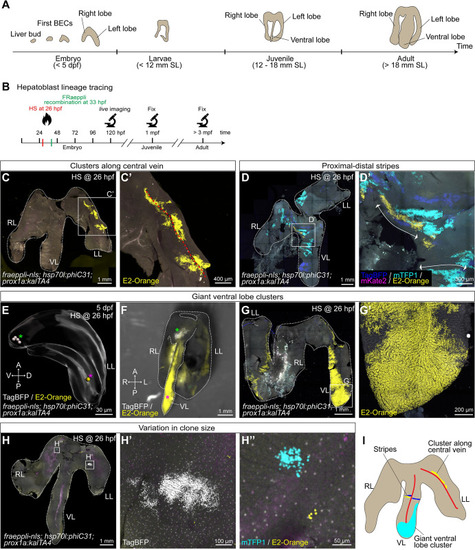
Lineage tracing reveals heterogeneous cluster topologies during postembryonic growth. ( |
|
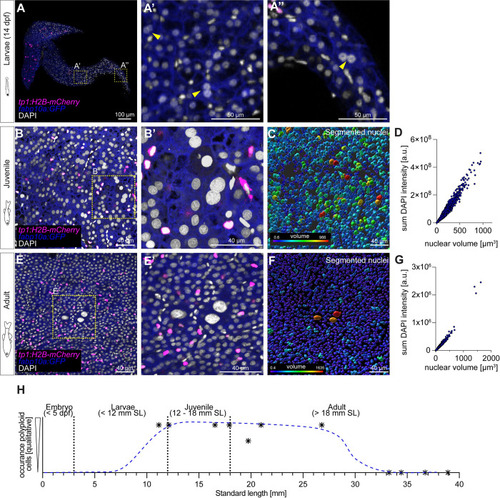
Polyploid cells appear transiently in hepatic postembryonic growth in zebrafish. ( |
|
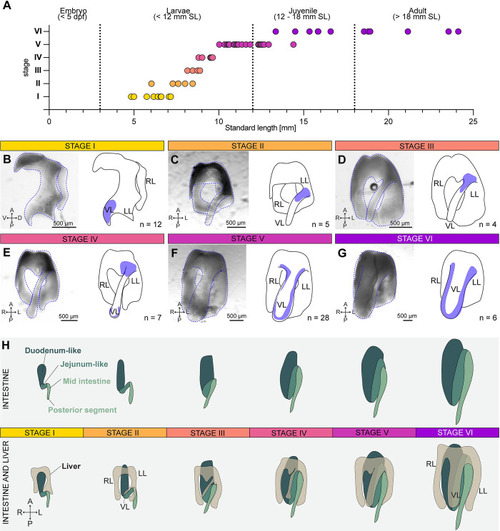
Ventral liver lobe formation during postembryonic growth. (A) The 6 steps of ventral liver lobe formation correlate with fish standard length (SL). The numerical values that were used to generate the graph can be found in |
|
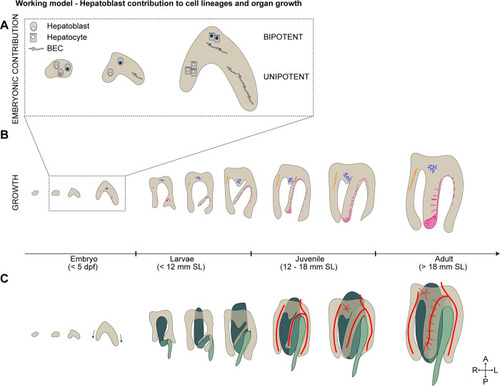
Working model of hepatoblast contribution to lineage decision and postembryonic growth. Schematics showing the current working models: ( |