- Title
-
Conservation of the insert-2 motif confers Rev1 from different species with an ability to disrupt G-quadruplexes and stimulate translesion DNA synthesis
- Authors
- Ketkar, A., Sewilam, R.S., McCrury, M.J., Hall, J.S., Bell, A., Paxton, B.C., Tripathi, S., Gunderson, J.E.C., Eoff, R.L.
- Source
- Full text @ RSC Chem Biol
|
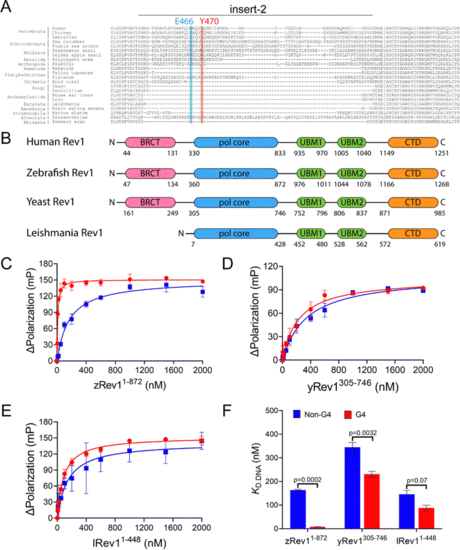
Comparison of G4 binding properties for Rev1 proteins from different species. (A) The sequence alignment of the insert-2 region for Rev1 proteins ranging from the SAR supergroup through vertebrata is shown. (B) A schematic illustration of domain arrangement and conservation is shown for the four Rev1 proteins used in this study. Binding curves for (C) zRev11-872 (D) yRev1305-746, and (E) lRev11-448 proteins with either the Myc 14/23 G4 DNA substrate (red circles) or non-G4 DNA control (blue squares) are shown. Protein was titrated into a solution containing either single-stranded (ss)-G4 DNA or ss-non-G4 DNA substrates at a concentration of 2 nM. The range of concentrations for the protein is indicated on the X-axis. The change in fluorescence polarization at each concentration was measured and plotted as a function of the protein concentration. (F) The changes in fluorescence polarization were fit to a quadratic equation to yield binding dissociation constants. The actual KD,DNA estimates are reported in Table 2. Values reported here represent the mean (± std. dev.) for three independent replicates. The reported p-values were calculated by using an unpaired Student's t-test with Welch's correction. |
|
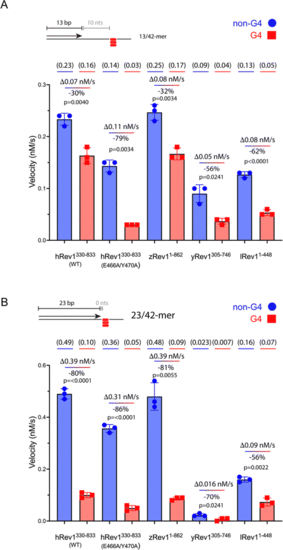
Rev1 catalysis is sensitive to G4 positioning relative to the primer terminus. (A) Single-nucleotide insertion experiments were performed with hRev1 (WT and EY mutant), zRev1, yRev1, and lRev1. A 13-mer primer was annealed to a 42-mer template strand containing either a non-G4 control sequence or the Myc 14/23 G4 motif. The initial rate of dCMP insertion was measured (nM s−1) and plotted for each enzyme. The absolute value for the rate of product formation is shown in parentheses above each data column. The absolute difference between the non-G4 and G4 DNA substrates, along with normalized change in activity for the G4 substrate, is noted for each enzyme. (B) Single-nucleotide insertion experiments were performed with hRev1 (WT and EY mutant), zRev1, yRev1, and lRev1. A 23-mer primer was annealed to a 42-mer template strand containing either a non-G4 control sequence or the Myc 14/23 G4 motif. The initial rate of dCMP insertion was measured (nM s−1) and plotted for each enzyme. The absolute value for the rate of product formation is shown in parentheses above each data column. The absolute difference between the non-G4 and G4 DNA substrates, along with normalized change in activity for the G4 substrate, is noted for each enzyme. Results shown represent the mean (± std. dev.) for three independent replicates. In both panels, results are shown for non-G4 control DNA substrates (blue circles) and G4 DNA substrates (red squares). The percent decrease in activity was calculated for each enzyme by considering the non-G4 control to be 100% active compared to the G4 substrate. The reported p-values were calculated by using an unpaired Student's t-test with Welch's correction. |
|
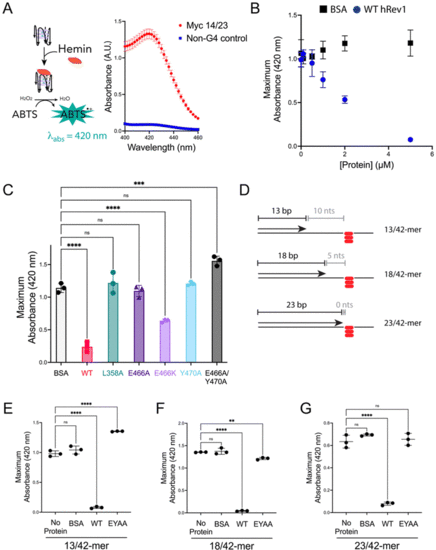
Mutations in insert-2 alter the ability of hRev1 to disrupt G4 DNA. (A) A cartoon schematic is shown depicting the DNAzyme-based assay monitoring G4 integrity. Briefly, binding of hemin to an intact G4 structure catalyzes oxidation of ABTS to a peroxidation product with an absorption maximum near 420 nm. Incubating the reaction mixture with a G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide produces a strong absorbance peak near 420 nm (red circles). Incubating the reaction mixture with a non-G4 ss-DNA (1 μM) oligonucleotide did not produce a detectable change in absorbance near 420 nm (blue squares). (B) The maximum absorbance at 420 nm was plotted as a function of protein concentration for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with either BSA (black squares) or WT hRev1330-833 (blue circles). (C) The maximum absorbance at 420 nm was measured for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with 5 μM of the indicated proteins. (D) Cartoon schematics are shown to depict the different primer-template DNA substrates used for G4 hemin assay results shown in panel (E–G). Please note that the 3′-OH is positioned 10, 5, or 0 nts away from the first tetrad-associated guanine for the 13-mer, 18-mer, and 23-mer primers, respectively. (E) The maximum absorbance at 420 nm was measured for reactions where 13/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (F) The maximum absorbance at 420 nm was measured for reactions where 18/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (G) The maximum absorbance at 420 nm was measured for reactions where 23/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. Results shown in all panels represent the mean (± std. dev.) for three replicates. p-Values in panels (C and E–G) were calculated using an ordinary one-way ANOVA with a Dunnett's multiple comparisons test where ** = p-value < 0.01, *** = p-value < 0.001, **** = p-value < 0.0001, and ns = not significant. |
|
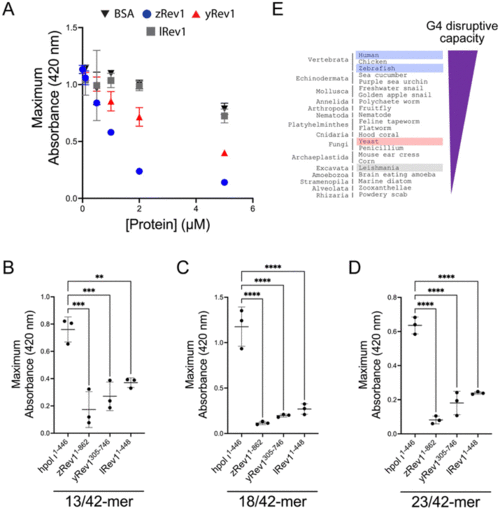
The presence of insert-2 increases the G4 disruptive capacity of Rev1. (A) The maximum absorbance at 420 nm was plotted as a function of protein concentration for reactions where G4-forming Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with either BSA (black inverted triangles), zRev11-862 (blue circles), yRev1305-746 (red triangles), or lRev11-448 (gray squares). (B) The maximum absorbance at 420 nm was measured for reactions where 13/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (C) The maximum absorbance at 420 nm was measured for reactions where 18/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (D) The maximum absorbance at 420 nm was measured for reactions where 23/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (E) Based on the results of the G4 hemin DNAzyme assay, the relative G4 disruptive capacity increased when moving across the tree of life from excavata through fungi to vertebrates. Results shown in panels (A–D) represent the mean (± std. dev.) for three replicates. p-Values in panels (B–D) were calculated using an ordinary one-way ANOVA with a Dunnett's multiple comparisons test where ** = p-value < 0.01, *** = p-value < 0.001, and **** = p-value < 0.0001. |
|
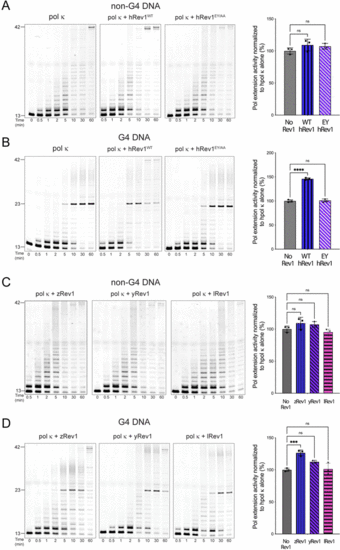
Insert-2 is important for stimulation of processive DNA synthesis across a G4 motif by pol κ. Polymerase extension assays were performed to measure the impact of Rev1 on DNA synthesis by a more processive TLS pol. Briefly, human pol κ (10 nM) was incubated with 13/42-mer primer-template DNA (200 nM) for 15 minutes before the reaction was initiated by the addition of solution that contained Rev1 (50 nM), MgCl2 (5 mM), and dNTP solution (0.2 mM total; 50 μM each for dATP, dCTP, dGTP, and dTTP). Pol extension was allowed to proceed at 37 °C before quenching at the indicated timepoints. Substrate and products were separated by PAGE using 14% (w/v) polyacrylamide gels with 7 M urea. (A) Pol extension assay results are shown for 13/42-mer non-G4 DNA where pol κ was incubated alone or in the presence of either WT hRev1 or the EY mutant. (B) Pol extension assay results are shown for 13/42-mer G4 DNA where pol κ was incubated alone or in the presence of either WT hRev1 or the EY mutant. (C) Pol extension assay results are shown for 13/42-mer non-G4 DNA where pol κ was incubated alone or in the presence of zRev1, yRev1, or lRev1. (D) Pol extension assay results are shown for 13/42-mer G4 DNA where pol κ was incubated alone or in the presence of zRev1, yRev1, or lRev1. Product formation for each reaction was quantified and normalized against the amount of product formed in the reaction with pol κ alone. The results are shown to the right of each set of gels and represent the mean (± std. dev.) of three independent replicates. Quantification of product formation by pol κ alone is re-plotted in each panel to allow more direct comparison with the reactions containing Rev1. p-Values were calculated using an ordinary one-way ANOVA with a Dunnett's multiple comparisons test where *** = p-value = 0.0008, **** = p-value < 0.0001, and ns = not significant. |
|
|