Fig. 2
- ID
- ZDB-FIG-231121-47
- Publication
- Ketkar et al., 2023 - Conservation of the insert-2 motif confers Rev1 from different species with an ability to disrupt G-quadruplexes and stimulate translesion DNA synthesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
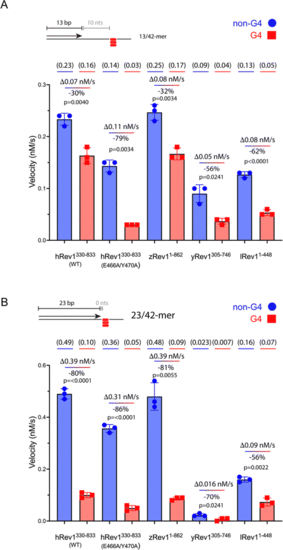
Rev1 catalysis is sensitive to G4 positioning relative to the primer terminus. (A) Single-nucleotide insertion experiments were performed with hRev1 (WT and EY mutant), zRev1, yRev1, and lRev1. A 13-mer primer was annealed to a 42-mer template strand containing either a non-G4 control sequence or the Myc 14/23 G4 motif. The initial rate of dCMP insertion was measured (nM s−1) and plotted for each enzyme. The absolute value for the rate of product formation is shown in parentheses above each data column. The absolute difference between the non-G4 and G4 DNA substrates, along with normalized change in activity for the G4 substrate, is noted for each enzyme. (B) Single-nucleotide insertion experiments were performed with hRev1 (WT and EY mutant), zRev1, yRev1, and lRev1. A 23-mer primer was annealed to a 42-mer template strand containing either a non-G4 control sequence or the Myc 14/23 G4 motif. The initial rate of dCMP insertion was measured (nM s−1) and plotted for each enzyme. The absolute value for the rate of product formation is shown in parentheses above each data column. The absolute difference between the non-G4 and G4 DNA substrates, along with normalized change in activity for the G4 substrate, is noted for each enzyme. Results shown represent the mean (± std. dev.) for three independent replicates. In both panels, results are shown for non-G4 control DNA substrates (blue circles) and G4 DNA substrates (red squares). The percent decrease in activity was calculated for each enzyme by considering the non-G4 control to be 100% active compared to the G4 substrate. The reported p-values were calculated by using an unpaired Student's t-test with Welch's correction. |