Fig. 3
- ID
- ZDB-FIG-231121-48
- Publication
- Ketkar et al., 2023 - Conservation of the insert-2 motif confers Rev1 from different species with an ability to disrupt G-quadruplexes and stimulate translesion DNA synthesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
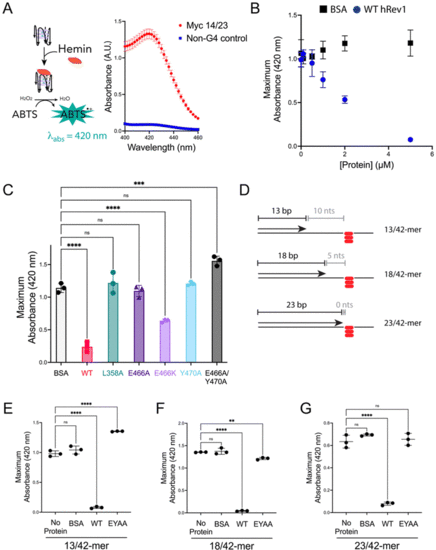
Mutations in insert-2 alter the ability of hRev1 to disrupt G4 DNA. (A) A cartoon schematic is shown depicting the DNAzyme-based assay monitoring G4 integrity. Briefly, binding of hemin to an intact G4 structure catalyzes oxidation of ABTS to a peroxidation product with an absorption maximum near 420 nm. Incubating the reaction mixture with a G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide produces a strong absorbance peak near 420 nm (red circles). Incubating the reaction mixture with a non-G4 ss-DNA (1 μM) oligonucleotide did not produce a detectable change in absorbance near 420 nm (blue squares). (B) The maximum absorbance at 420 nm was plotted as a function of protein concentration for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with either BSA (black squares) or WT hRev1330-833 (blue circles). (C) The maximum absorbance at 420 nm was measured for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with 5 μM of the indicated proteins. (D) Cartoon schematics are shown to depict the different primer-template DNA substrates used for G4 hemin assay results shown in panel (E–G). Please note that the 3′-OH is positioned 10, 5, or 0 nts away from the first tetrad-associated guanine for the 13-mer, 18-mer, and 23-mer primers, respectively. (E) The maximum absorbance at 420 nm was measured for reactions where 13/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (F) The maximum absorbance at 420 nm was measured for reactions where 18/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (G) The maximum absorbance at 420 nm was measured for reactions where 23/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. Results shown in all panels represent the mean (± std. dev.) for three replicates. p-Values in panels (C and E–G) were calculated using an ordinary one-way ANOVA with a Dunnett's multiple comparisons test where ** = p-value < 0.01, *** = p-value < 0.001, **** = p-value < 0.0001, and ns = not significant. |