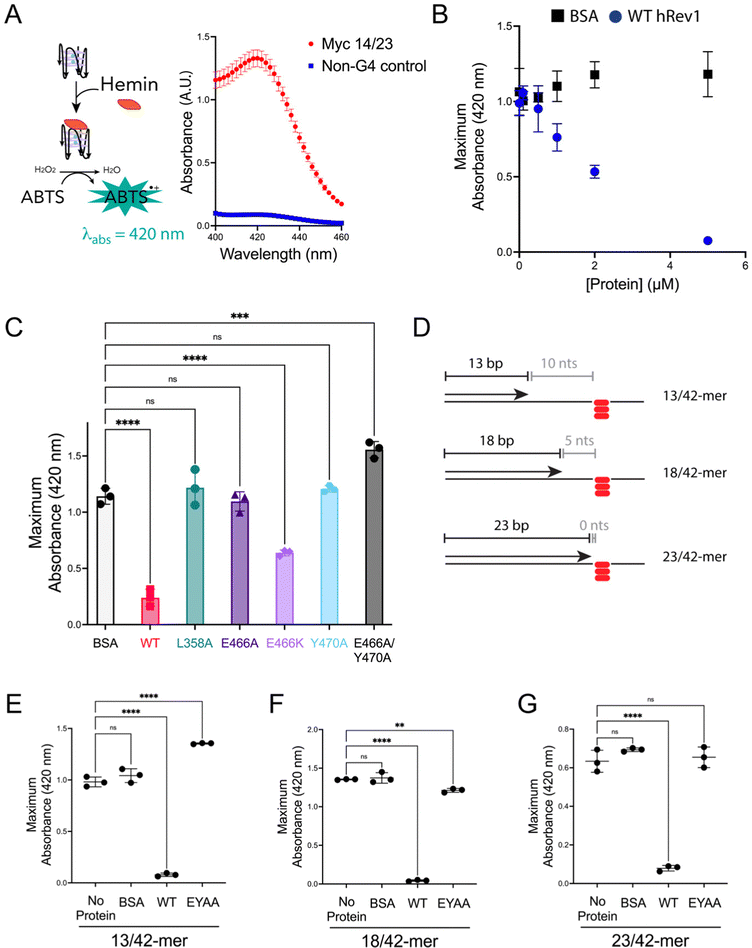
Fig. 3 Mutations in insert-2 alter the ability of hRev1 to disrupt G4 DNA. (A) A cartoon schematic is shown depicting the DNAzyme-based assay monitoring G4 integrity. Briefly, binding of hemin to an intact G4 structure catalyzes oxidation of ABTS to a peroxidation product with an absorption maximum near 420 nm. Incubating the reaction mixture with a G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide produces a strong absorbance peak near 420 nm (red circles). Incubating the reaction mixture with a non-G4 ss-DNA (1 μM) oligonucleotide did not produce a detectable change in absorbance near 420 nm (blue squares). (B) The maximum absorbance at 420 nm was plotted as a function of protein concentration for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with either BSA (black squares) or WT hRev1330-833 (blue circles). (C) The maximum absorbance at 420 nm was measured for reactions where G4-forming 29-mer Myc 14/23 ss-DNA (1 μM) oligonucleotide was incubated with 5 μM of the indicated proteins. (D) Cartoon schematics are shown to depict the different primer-template DNA substrates used for G4 hemin assay results shown in panel (E–G). Please note that the 3′-OH is positioned 10, 5, or 0 nts away from the first tetrad-associated guanine for the 13-mer, 18-mer, and 23-mer primers, respectively. (E) The maximum absorbance at 420 nm was measured for reactions where 13/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (F) The maximum absorbance at 420 nm was measured for reactions where 18/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. (G) The maximum absorbance at 420 nm was measured for reactions where 23/42-mer DNA with the Myc 14/23 G4 motif in the template strand (1 μM) was incubated with 5 μM of the indicated proteins. Results shown in all panels represent the mean (± std. dev.) for three replicates. p-Values in panels (C and E–G) were calculated using an ordinary one-way ANOVA with a Dunnett's multiple comparisons test where ** = p-value < 0.01, *** = p-value < 0.001, **** = p-value < 0.0001, and ns = not significant.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ RSC Chem Biol