- Title
-
Genome-wide screening in pluripotent cells identifies Mtf1 as a suppressor of mutant huntingtin toxicity
- Authors
- Ferlazzo, G.M., Gambetta, A.M., Amato, S., Cannizzaro, N., Angiolillo, S., Arboit, M., Diamante, L., Carbognin, E., Romani, P., La Torre, F., Galimberti, E., Pflug, F., Luoni, M., Giannelli, S., Pepe, G., Capocci, L., Di Pardo, A., Vanzani, P., Zennaro, L., Broccoli, V., Leeb, M., Moro, E., Maglione, V., Martello, G.
- Source
- Full text @ Nat. Commun.
|
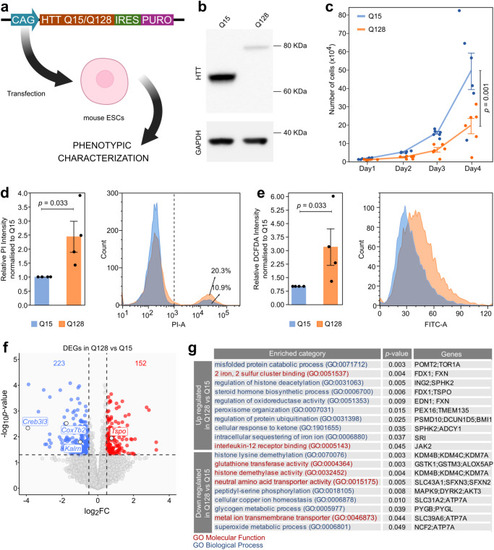
Establishment and characterisation of mHTT-expressing mouse ESCs. |
|
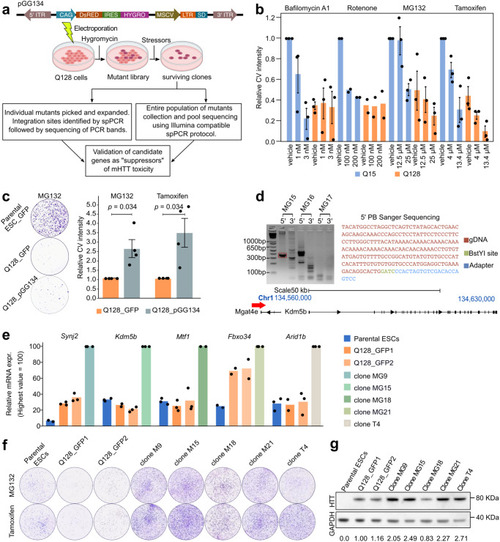
A gain-of-function screen for suppressors of mHTT toxicity. |
|
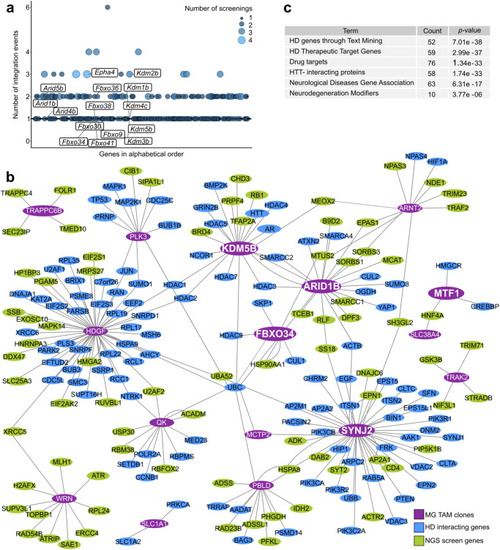
Network analysis of candidate suppressors of mHTT toxicity. |
|
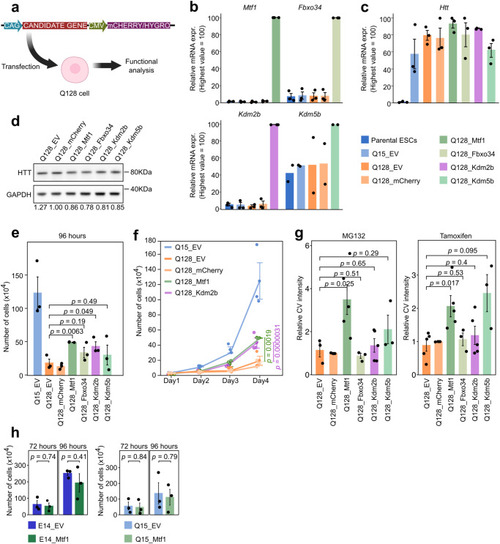
Secondary validation of mHTT suppressors. |
|
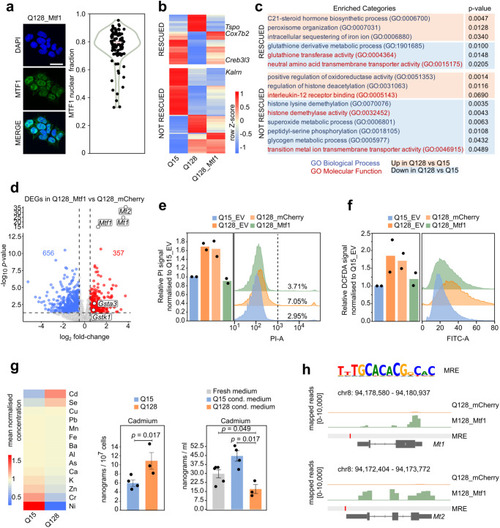
Mtf1 regulates HD-related processes |
|
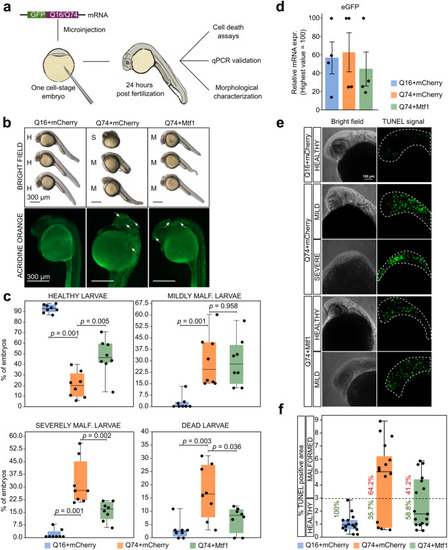
Mtf1 counteracts mHTT effects in zebrafish. |
|
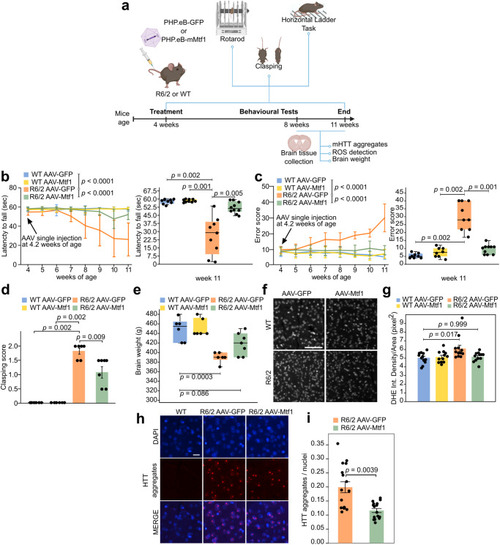
AAV-vector delivery of Mtf1 alleviates motor deficit in R6/2 mice. |
|
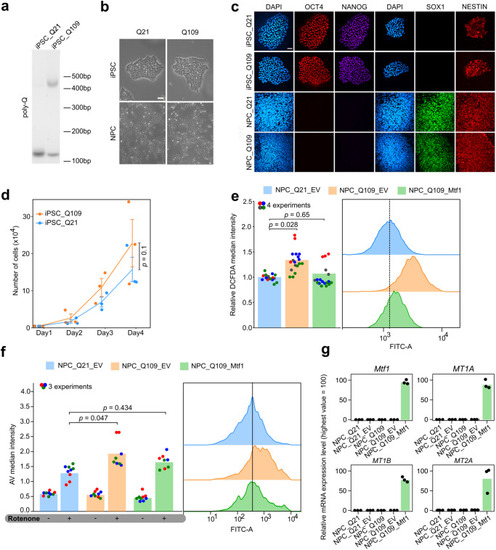
MTF1 rescues mHTT-dependent alterations in human NPCs. |