- Title
-
Intrinsic epigenetic control of angiogenesis in induced pluripotent stem cell-derived endothelium regulates vascular regeneration
- Authors
- Macklin, B.L., Lin, Y.Y., Emmerich, K., Wisniewski, E., Polster, B.M., Konstantopoulos, K., Mumm, J.S., Gerecht, S.
- Source
- Full text @ NPJ Regen Med
|
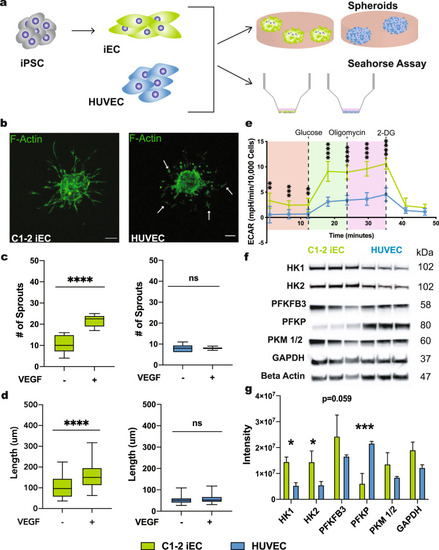
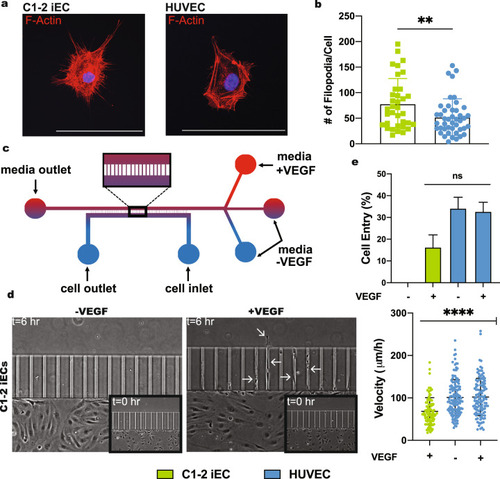
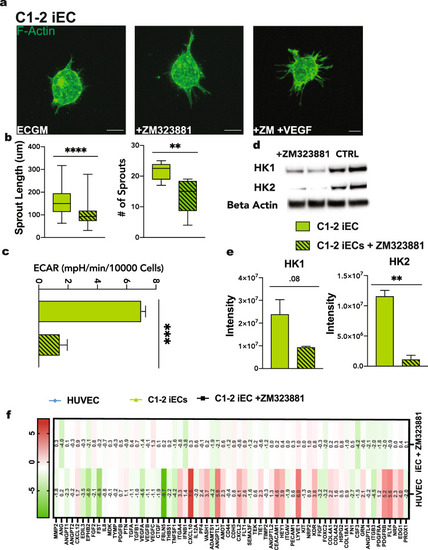
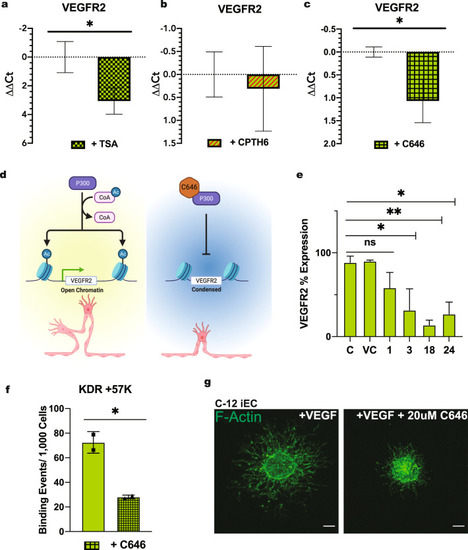
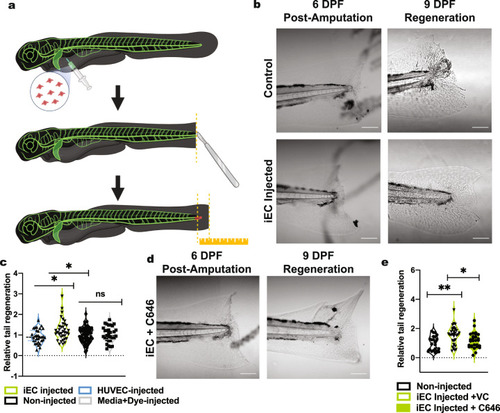
iECs derived from the C1-2 hiPSC line were analyzed for sprouting abilities. |
|
|
|
|
|
|
|
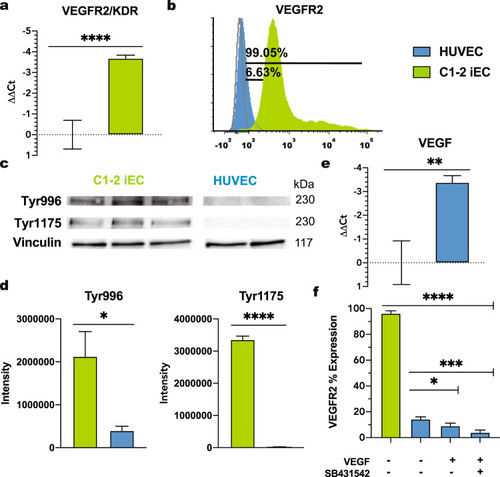
RT-qPCR for VEGFR2 in C1-2 iECs treated with |
|
|