- Title
-
Lifelong single-cell profiling of cranial neural crest diversification in zebrafish
- Authors
- Fabian, P., Tseng, K.C., Thiruppathy, M., Arata, C., Chen, H.J., Smeeton, J., Nelson, N., Crump, J.G.
- Source
- Full text @ Nat. Commun.
|
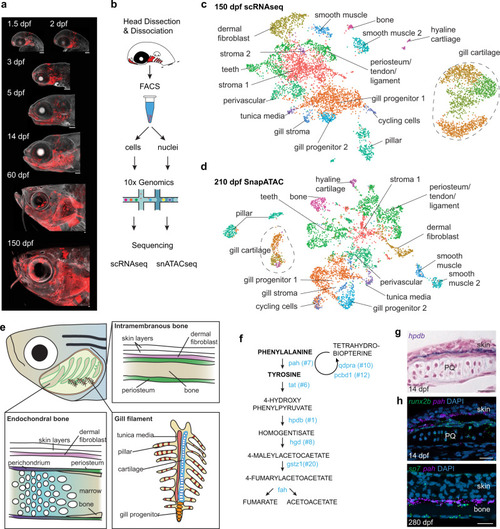
a Sox10:Cre; actab2:loxP-BFP-STOP-loxP-dsRed labeling of CNCCs (red) in zebrafish heads across 7 stages. b Scheme of cell or nuclei dissociation, fluorescence-activated cell sorting (FACS), and processing on the 10X Genomics platform for sequencing. c, d UMAPs of scRNAseq and SnapATAC datasets at adult stages. e Diagram of an adult zebrafish head cut out to show major cell types of the interior skeletal elements and gill system. f Pathway for Phe and Tyr breakdown. 7 of 8 genes encoding catabolic enzymes are in the top 20 selectively enriched genes for the dermal fibroblast cluster (numbers show rank). g Section colorimetric in situ hybridization for hpdb RNA shows expression in the dermis between the skin and palatoquadrate (PQ) endochondral bone. h Section RNAscope in situ hybridizations show pah expression in dermal fibroblasts between the runx2b+ periosteum of PQ and skin at 14 dpf, and between sp7+ osteoblasts of intramembranous bone and skin at 280 dpf. DAPI labels nuclei in blue. Scale bars, 100 μm (a), 20 μm (g, h). EXPRESSION / LABELING:
|
|
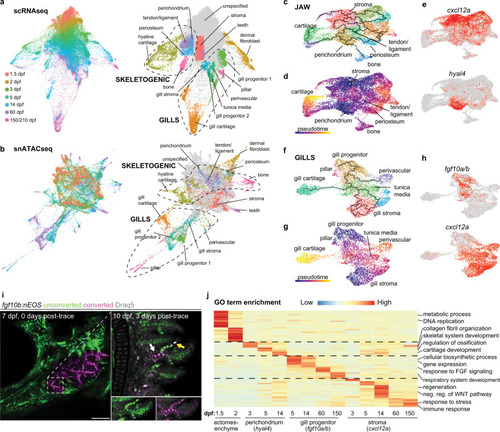
a, b STITCH plots connect individual scRNAseq and snATACseq datasets across the zebrafish lifetime. Cell type annotations show divergence of CNCC ectomesenchyme into skeletogenic, gill, dermal fibroblast, and stromal branches. c–e Pseudotime analysis using Monocle3 of the jaw skeletogenic subset (combined 5 and 14 dpf) shows a cxcl12a+ stromal branch, and a hyal4+ branch connected to cartilage, bone, periosteum, and tendon and ligament. f–h Pseudotime analysis of gill subsets (combined 5 and 14 dpf) shows a cxcl12a+ stromal branch, and a fgf10a/b+ branch connected to gill pillar, tunica media, and perivascular cells, as well as a distinct type of gill cartilage. i Following UV-mediated photoconversion of fgf10b:nEOS from green to magenta in a subset of filaments, re-imaging 3 days later revealed contribution of converted cells to gill chondrocytes (white arrow) and pillar cells (yellow arrow, white signal reflects mixture of previously converted magenta and continued expression of new unconverted green fgf10b:nEOS) (n = 3). Draq5 labels nuclei in gray. At 10 dpf, below insets show the region designated by the arrows for individual green and magenta channels with Draq5. j Gene ontology (GO) terms of biological processes for the respective clusters at the stages indicated. Heatmap reflects the negative log of the adjusted p value of one-sided Fisher’s exact test. Ectomesenchyme is defined as the average of all cells at 1.5 and 2 dpf stages. Scale bar, 100 μm. PHENOTYPE:
|
|
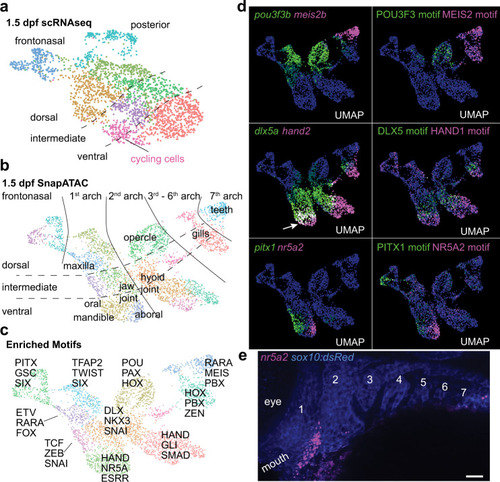
a, b, UMAPs at 1.5 dpf generated by scRNAseq versus integration of scRNAseq and snATACseq datasets using SnapATAC. SnapATAC outperforms scRNAseq in resolving dorsoventral (vertical), anterior-posterior (horizontal), and major arch landmarks including a previously unappreciated oral-aboral axis in zebrafish. c Select enriched transcription factor binding motifs for each cluster. d Gene body activities for transcription factors and their corresponding DNA-binding motifs reveal tight correspondence with published expression patterns, including zebrafish-specific overlap of dlx5a and hand2 in the mandibular arch (arrow). e Fluorescent in situ hybridization shows restricted expression of nr5a2 in the aboral domain of the mandibular arch as predicted by SnapATAC. sox10:dsRed labels CNCCs of the arches in blue (numbered, anti-dsRed antibody stain). Scale bar, 20 μm. |
|
a UMAP of 14 dpf SnapATAC data shows cell clusters for which top accessible peak modules were calculated for Constellations analysis. b Constellations analysis involves mapping of cluster-specific chromatin accessibility from 14 dpf back to earlier stages and then plotting relatedness of mapped accessibility in two dimensions. Diffuse refers to a stage when cluster-specific chromatin accessibility does not map to a discrete portion of UMAP space, and skewed when it does (see “Methods” for details). Groups of related cell types are color-coded. c Graphical representation from the Constellations analysis of when chromatin accessibility of major cell types first shows a skewed distribution in UMAP space, suggestive of establishment of competency. PHENOTYPE:
|
|
a Mapping onto the Constellations plot of transcription factors with correlated gene body activity and DNA-binding motif enrichment in specific clusters. Sizes of circles denote correlation of peak module mapping to motifs, and red color to gene body activities. b Top peak modules for hyaline cartilage and dermal fibroblast 1 clusters at 14 dpf mapped onto 14 dpf and 1.5 dpf SnapATAC UMAPs. c Summed gene body activities of foxc1a, foxc1b, foxf1, foxf2a, and foxf2b and summed FOXC1, FOXF1, and FOXF2 motifs at 1.5 dpf correlate with cartilage peak mapping, and lef1 gene body activity and LEF1 motif with dermal fibroblast peak mapping. d, e Retrograde cartilage accessibility mapping at 1.5 dpf allows predictions of the arch origins of the individual cartilaginous elements of the week-old skeleton: ceratobranchials (CBs), ceratohyal (CH), ethmoid (ET), hyomandibula (HM), Meckel’s (M), palatoquadrate (PQ), symplectic (SY), and trabecula (TR). f Mapping of the top peak module for 14 dpf gill progenitor clusters onto 5 dpf SnapATAC UMAP shows correlation with gata3 gene body activity and GATA3 motif. |
|
a, b Genome tracks show chromatin accessibility for cell clusters from snATACseq data at 14 dpf. Gray shading shows gill-specific accessible regions near the gata3 and ucmaa genes. Chromosome positions refer to the GRCz11 genome assembly. c–f gata3-P1:GFP drives expression in the posterior gill-forming arches at 3 dpf, the gill filament system at 14 dpf, and gill progenitors at the tips of primary filaments at 60 dpf, as well as some pillar cells (arrowhead) and gill chondrocytes (arrow) near the growing tips. sox10:dsRed labels cartilage and Draq5 nuclei. f ucmaa-P1:GFP drives highly restricted expression in sox10:dsRed+ gill chondrocytes (boxed region shown in merged and single channels to the right) but not hyaline cartilage (top left). g Model shows the initiation of gata3 expression in the posterior gill-forming arches through the gata3-P1 enhancer, maintenance of gata3 in gill progenitors at the tips of growing filaments, and expression of ucmaa in gill cartilage through the ucmaa-P1 enhancer. Both gata3-P1 and ucmaa-P1 contain predicted Gata3 binding sites. Scale bars, 100 μm (c, d, f), 20 μm (e). EXPRESSION / LABELING:
PHENOTYPE:
|
|
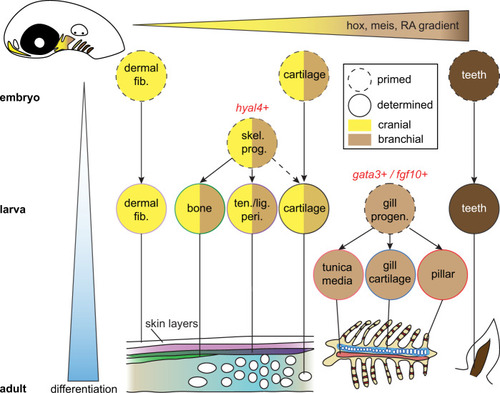
Cell types progressively emerge at distinct positions along the anterior-posterior axis of the developing head. Time is represented along the y-axis from young (white) to old (blue). Positional information is represented along the x-axis from anterior (yellow) to posterior (brown). Cell types are denoted by circles, with skeletal progenitors distinguished by hyal4 expression and gill progenitors by gata3 and fgf10 gene expression. Mature cell types are schematized at the bottom. |