- Title
-
Neuropeptide VF neurons promote sleep via the serotonergic raphe
- Authors
- Lee, D.A., Oikonomou, G., Cammidge, T., Andreev, A., Hong, Y., Hurley, H., Prober, D.A.
- Source
- Full text @ Elife
|
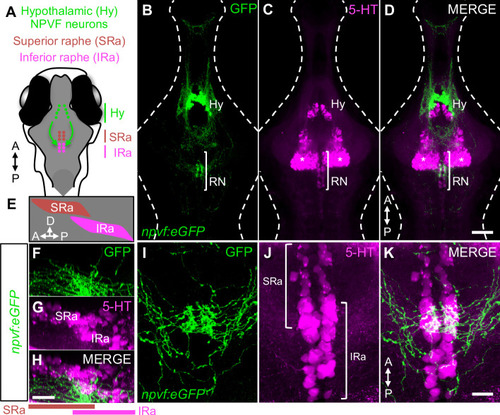
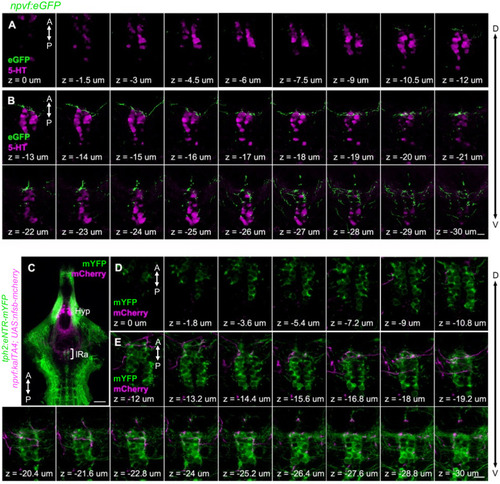
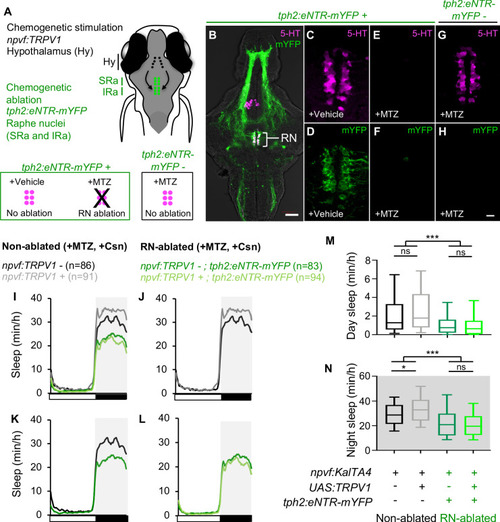
Hypothalamic NPVF neurons project to the serotonergic IRa. (A,E) Schematic: 6-dpf zebrafish brain showing location of hypothalamic (Hy) NPVF neurons (green), and the serotonergic superior raphe (SRa, red) and inferior raphe (IRa, magenta). A, anterior; P, posterior; D, dorsal. (B–D) Maximum intensity projection of a brain from a 6-dpf Tg(npvf:eGFP) animal (78 μm thick). npvf-expressing neurons in the hypothalamus project to the serotonergic raphe nuclei (RN) in the hindbrain (bracket). 5-HT immunohistochemistry labels the RN (bracket), as well as serotonergic populations in the ventral hypothalamus (asterisks) and pretectum. The bracketed region in (B–D) is shown at higher magnification in (I–K) as a maximum intensity projection (50.5 μm thick), with a sagittal view shown in (F–H). Single optical sections are shown in Figure 1—figure supplement 1. Scale: 50 μm (B–D), 20 μm (F–H), and 10 μm (I–K). |
|
( |
|
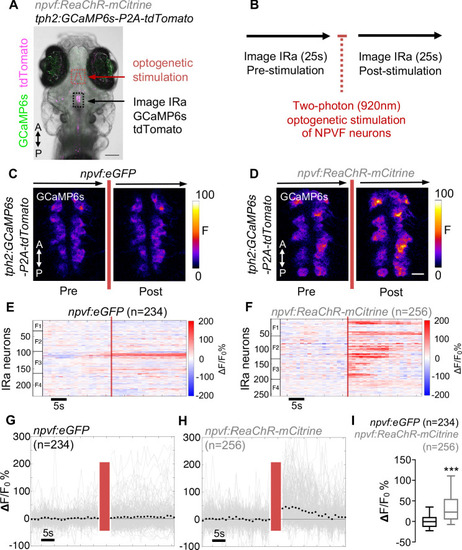
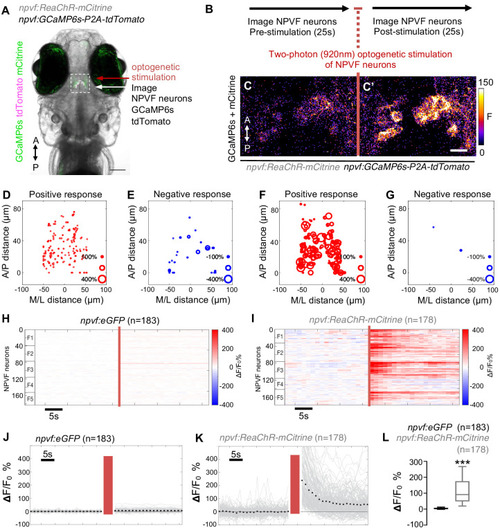
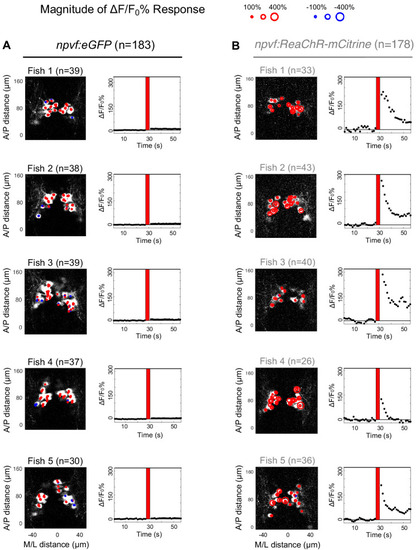
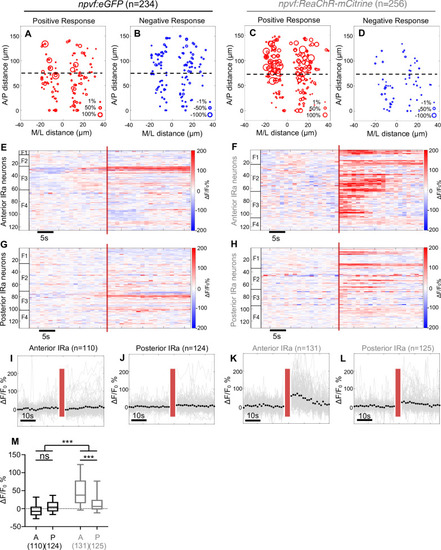
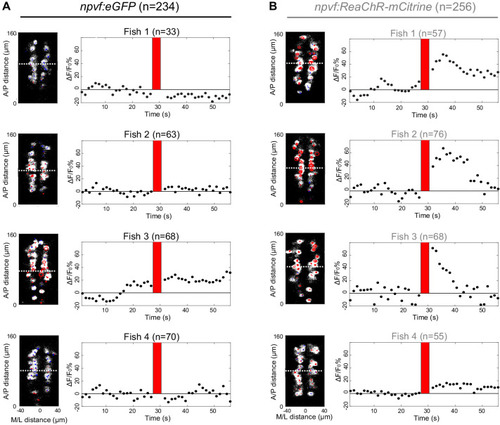
Optogenetic stimulation of NPVF neurons activates serotonergic IRa neurons. 6-dpf Tg(npvf:ReaChR-mCitrine); Tg(tph2:GCaMP6s-P2A-tdTomato) and Tg(npvf:eGFP); Tg(tph2:GCaMP6s-P2A-tdTomato) control animals were analyzed for GCaMP6s/tdTomato fluorescence levels in IRa neurons before and after optogenetic stimulation of NPVF neurons. (A) Example of a 6-dpf Tg(npvf:ReaChR-mCitrine); Tg(tph2:GCaMP6s-P2A-tdTomato) animal showing GCaMP6s and tdTomato fluorescence in IRa neurons with overlayed NPVF neurons (red circles, different z-plane) indicating region of optogenetic stimulation (red box). Black box indicates region where IRa neurons were imaged and analyzed in subsequent panels. Green and magenta signals in the eyes are due to autofluorescence. (B) Normalized GCaMP6s fluorescence in the IRa was first recorded for 25 s (20 frames). NPVF neurons were then optogentically stimulated for 3.7 s (red line), and normalized GCaMP6s fluorescence in the IRa was then immediately imaged again for 25 s. (C,D) Average GCaMP6s fluorescence in IRa neurons for 10 imaging frames before (Pre) and after (Post) optogenetic stimulation of NPVF neurons in Tg(npvf:eGFP); Tg(tph2:GCaMP6s-P2A-tdTomato) (C) and Tg(npvf:ReaChR-mCitrine); Tg(tph2:GCaMP6s-P2A-tdTomato) (D) animals. (E–H) Normalized GCaMP6s fluorescence of individual IRa neurons shown as heat maps (E,F), in which each horizontal line represents an IRa neuron, and as line graphs (G,H) showing individual (gray lines) and mean (dotted line) IRa neuron responses before and after optogenetic stimulation (red lines) in Tg(npvf:eGFP); Tg(tph2:GCaMP6s-P2A-tdTomato) control (E,G) and Tg(npvf:ReaChR-mCitrine); Tg(tph2:GCaMP6s-P2A-tdTomato) (F,H) animals. F1-F4 in (E,F) indicate neurons from four different fish. (I) Box plot of normalized GCaMP6s ΔF/F0 values from the average of the first 10 imaging frames post-stimulation of each neuron for Tg(tph2:GCaMP6s-P2A-tdTomato) animals that also contain either a Tg(npvf:eGFP) (black) or Tg(npvf:ReaChR-mCitrine) (gray) transgene. n = number of neurons quantified from four animals of each genotype. ***p<0.001, Mann-Whitney test. Scale: 100 μm (A), 10 μm (D). |
|
6-dpf |
|
6-dpf |
|
( |
|
6-dpf |
|
( |
|
( |
|
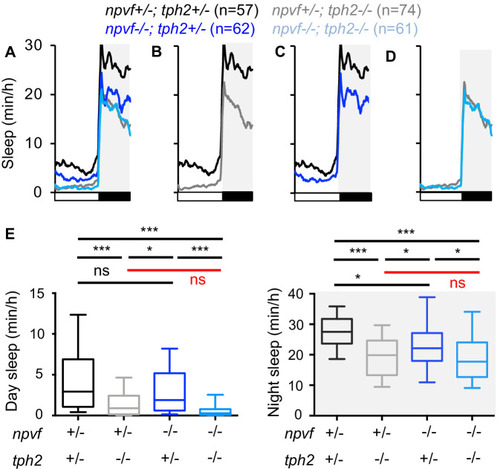
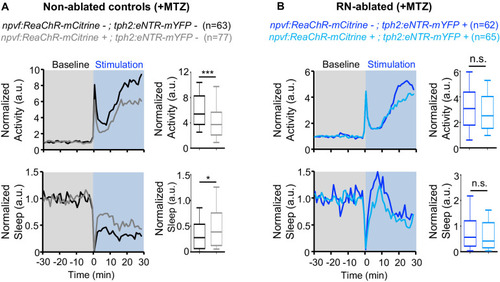
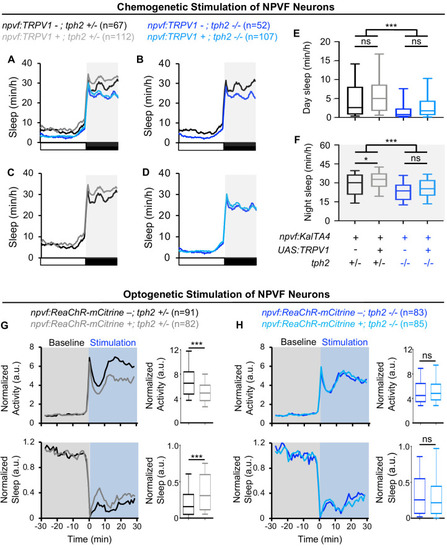
Normalized locomotor activity (top) and sleep (bottom) of 5-dpf Tg(npvf:ReaChR-mCitrine) (gray and light blue) and non-transgenic sibling control (black and blue) animals before (Baseline) and during blue light exposure (Stimulation) in Tg(tph2:eNTR-mYFP) negative (A) or positive (B) siblings. Because the animals see the blue light, they exhibit a brief startle at light onset that is excluded from analysis, followed by a gradual increase in activity that plateaus after ~15 min. Box plots quantify locomotor activity and sleep for each animal during optogenetic stimulation normalized to the baseline of all animals of the same genotype. n = number of animals. ns p>0.05, *p<0.05, ***p<0.005, Mann-Whitney test. |
|
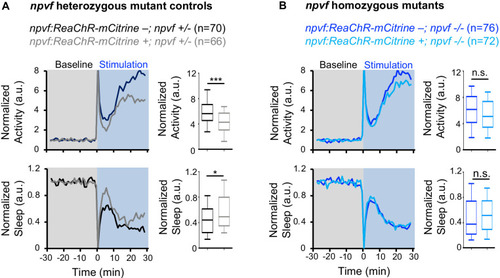
Normalized locomotor activity (top) and sleep (bottom) of 5-dpf |
|
( |