- Title
-
Hcfc1a regulates neural precursor proliferation and asxl1 expression in the developing brain
- Authors
- Castro, V.L., Reyes, J.F., Reyes-Nava, N.G., Paz, D., Quintana, A.M.
- Source
- Full text @ BMC Neurosci.
|
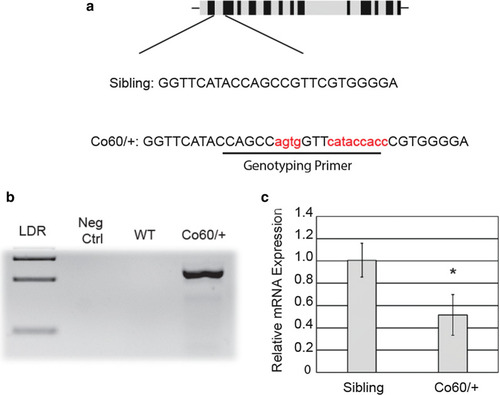
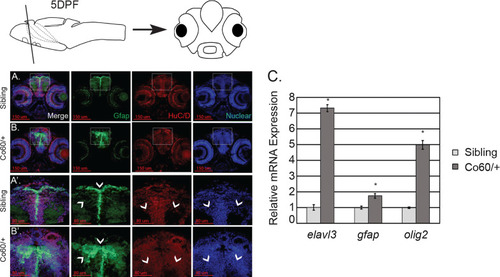
Haploinsufficiency of the |
|
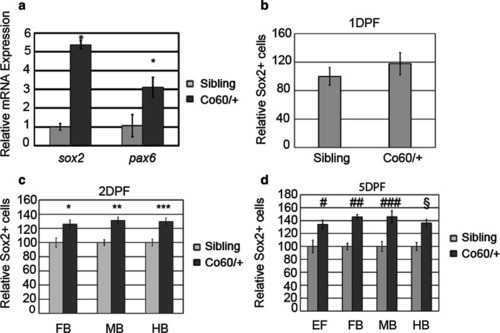
The |
|
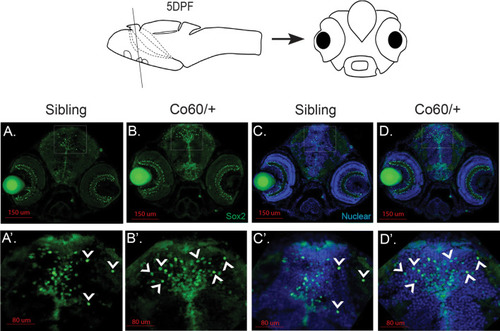
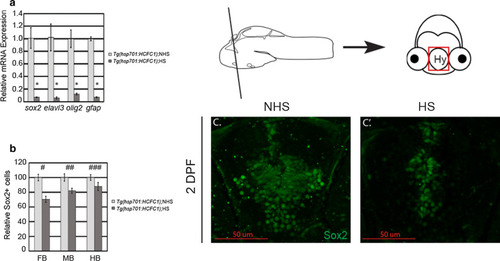
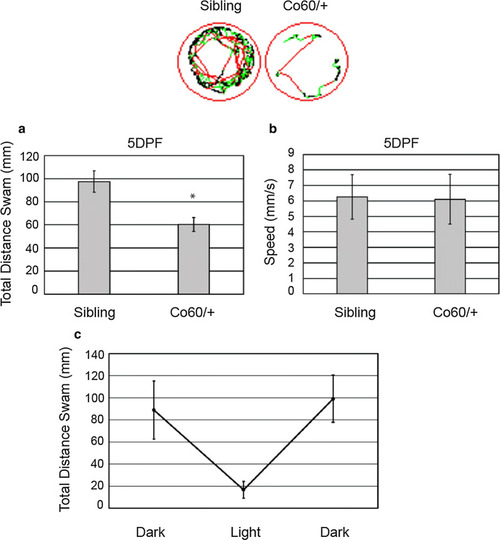
Increased Sox2+ cells are present in the |
|
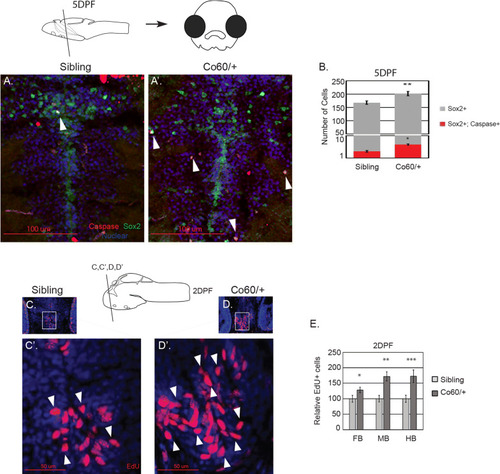
The |
|
The |
|
Over expression of HCFC1 decreases NPC number and differentiation. |
|
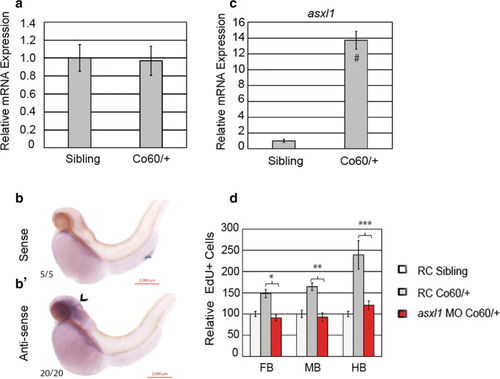
The |
|
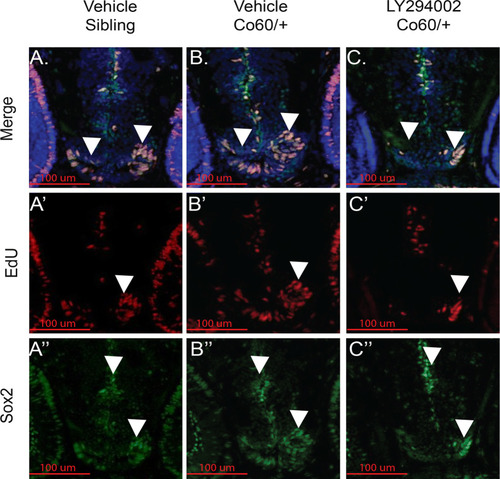
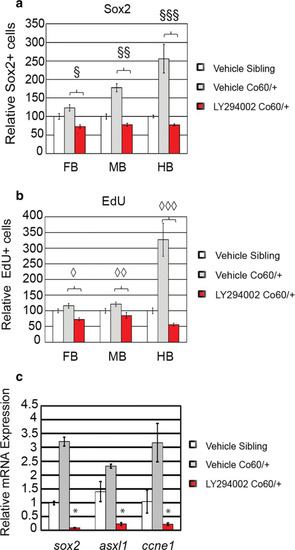
Inhibition of Asxl1 activity restores the NPC deficits in the PHENOTYPE:
|
|
Quantification of the number of NPCs after inhibition of Asxl1 activity. |
|
The PHENOTYPE:
|