- Title
-
A network-based approach to identify deregulated pathways and drug effects in metabolic syndrome
- Authors
- Misselbeck, K., Parolo, S., Lorenzini, F., Savoca, V., Leonardelli, L., Bora, P., Morine, M.J., Mione, M.C., Domenici, E., Priami, C.
- Source
- Full text @ Nat. Commun.
|
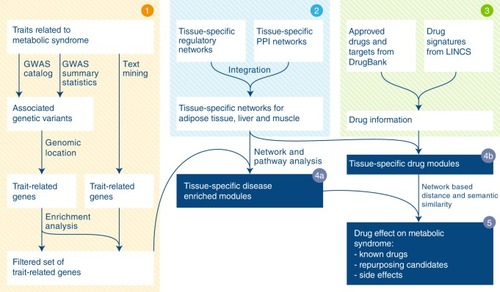
Schematic illustration of the computational framework. Step 1 MetSyn-related genes are identified by combining GWAS results and literature findings followed by a filtering step based on gene-set enrichment analysis. Step 2 Tissue-specific networks are constructed by integrating transcriptional regulatory networks from |
|
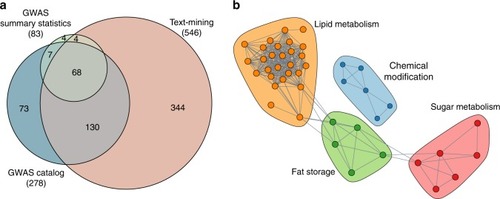
Identification of MetSyn-related genes. |
|
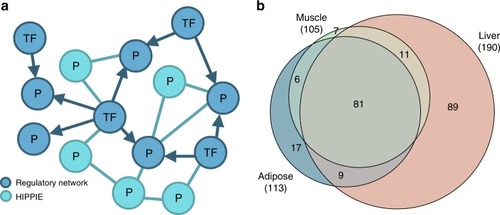
Network construction and disease module identification. |
|
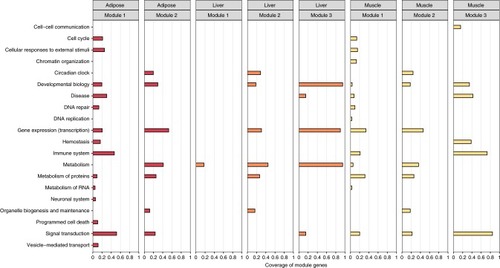
Functional annotation of the disease modules. For each network and disease module the coverage of module genes by significantly enriched Reactome pathways, grouped according to the TopLevel pathway classification, is presented. See also Supplementary Data |
|
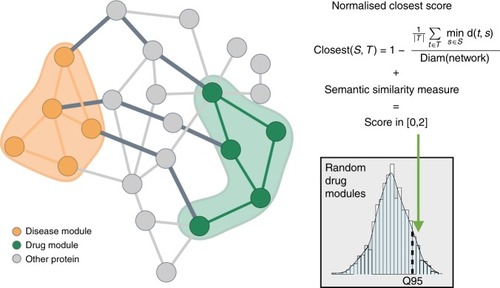
Illustration of the score calculation to evaluate the drug-disease interplay. The score combines network distances and functional similarity between proteins in the drug module (green nodes) and proteins in the disease module (orange nodes). The network score assesses the shortest path lengths connecting each protein of the drug module to the nearest protein in the disease module (dark gray edges) while the semantic similarity measure evaluates the functional similarity between the modules. To test the score significance, the distance between drug and disease modules is compared to a reference distribution of scores computed with drug modules randomly chosen from the network |
|
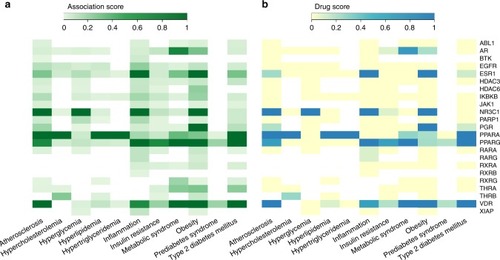
Association between the active drug targets in the adipose network and MetSyn-related traits based on the scores provided by OpenTargets. |
|
|
|
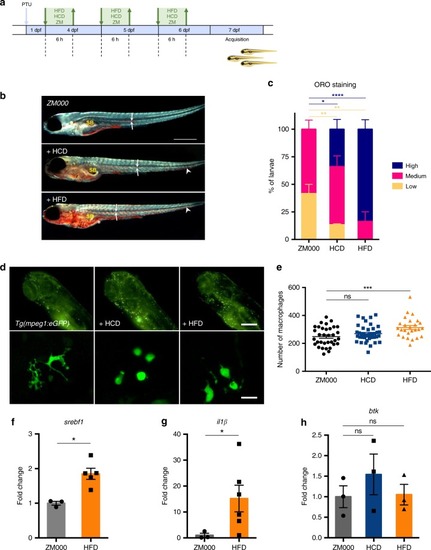
High fat diet induces lipid and macrophage accumulation in zebrafish. |
|
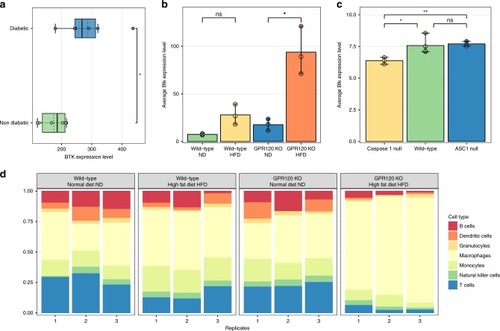
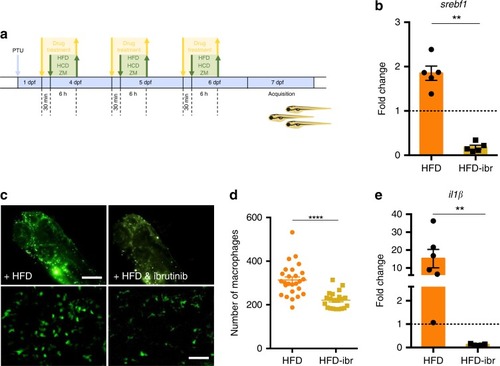
High fat diet effects can be prevented by ibrutinib treatment. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|