- Title
-
ADP-dependent glucokinase regulates energy metabolism via ER-localized glucose sensing
- Authors
- Imle, R., Wang, B.T., Stützenberger, N., Birkenhagen, J., Tandon, A., Carl, M., Himmelreich, N., Thiel, C., Gröne, H.J., Poschet, G., Völkers, M., Gülow, K., Schröder, A., Carillo, S., Mittermayr, S., Bones, J., Kamiński, M.M., Kölker, S., Sauer, S.W.
- Source
- Full text @ Sci. Rep.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
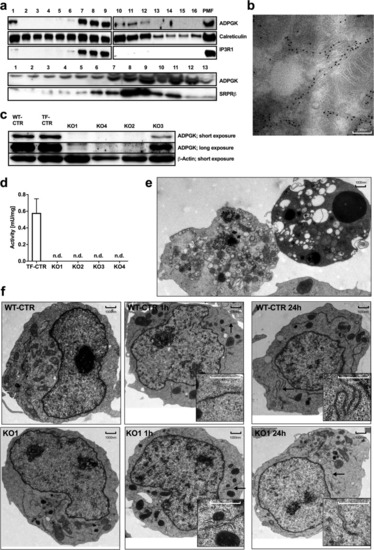
ADPGK is localized in ER lumen and important for ER biogenesis. |
|
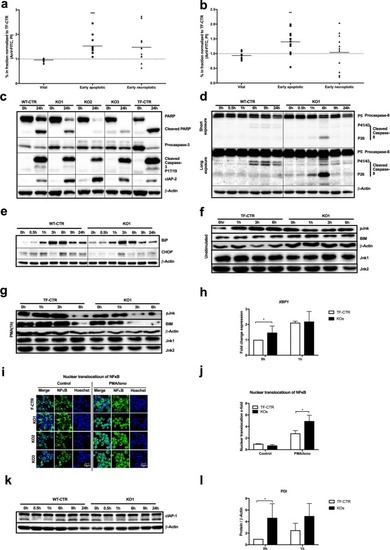
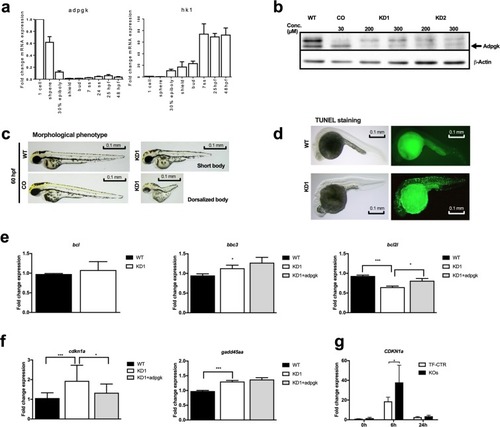
ADPGK knockout leads to apoptosis and ER stress. |
|
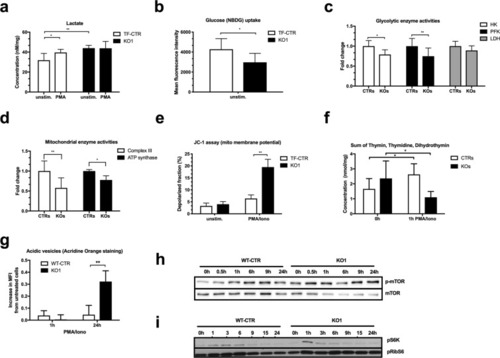
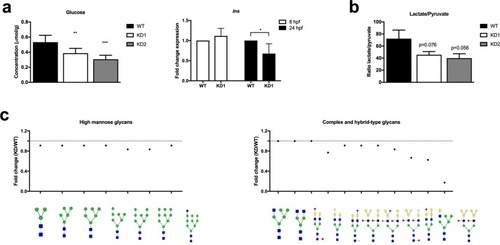
ADPGK regulates energy metabolism. ( |
|
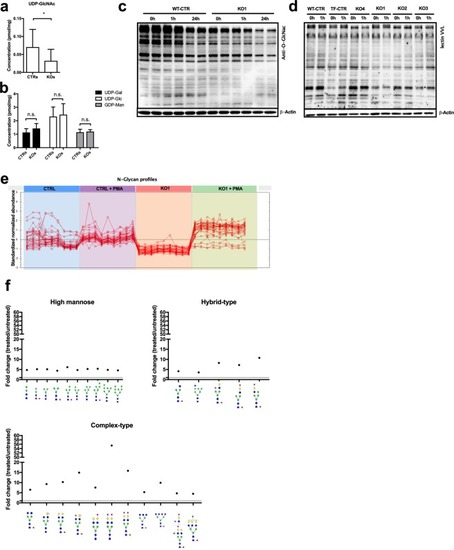
ADPGK finetunes glucose flux into O- and N-glycosylation. |
|
Knockdown of Adpgk in embryonic zebrafish results in aberrant body axis development and cell death. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Adpgk regulates glucose metabolism in zebrafish embryos. ( EXPRESSION / LABELING:
PHENOTYPE:
|