- Title
-
Gene expression and functional analysis of zebrafish larval fin fold regeneration
- Authors
- Yoshinari, N., Ishida, T., Kudo, A., and Kawakami, A.
- Source
- Full text @ Dev. Biol.
|
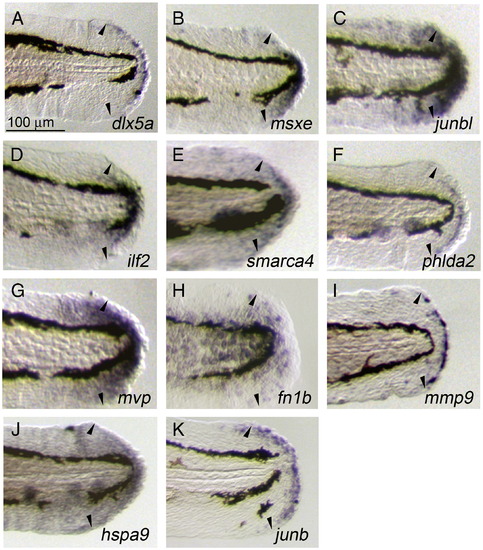
Induced and localized expression of regeneration-related transcripts during larval regeneration. (A–K) Whole-mount ISH analysis showing the respective gene expression at 1 dpa during larval regeneration. The expressions of dlx5a (A) and junb (K) were seen in the epithelial cells at the stump that appear to correspond to the wound epithelium, whereas the expressions of msxe (B), junbl (C), ilf2 (D), smarca4 (E), mvp (G), and hspa9 (J) were in the mesenchymal regions containing the blastema-like proliferating cells. In addition to these characterized cell populations, the expression of phlda2 was seen in a sharp line of cells between the wound epithelium and mesenchyme (F), mmp9 in a population of epithelial cells with an irregular distribution (I), and fn1b in a broad epithelial region (H). For all genes, expression was not detected in uncut fin folds at the same developmental stage (data not shown). Arrowheads indicate the edges of amputated areas. The same magnifications for panels A–K (scale bar in panel A). EXPRESSION / LABELING:
|
|
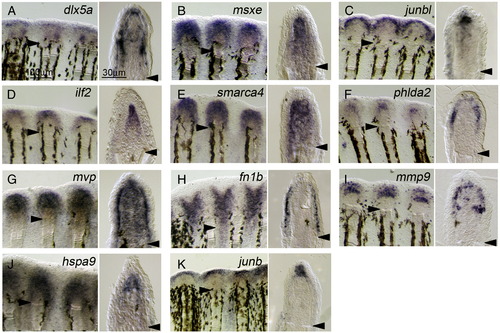
Localized expression of regeneration-related transcripts during adult regeneration. (A–K) ISH analysis showing the respective gene expression at 2 dpa of adult regeneration. Respective gene expressions were detected in whole-mount preparations (left panels), and their cellular localizations were assessed in sections (right panels). The expression of dlx5a (A) had 3 domains: the basal wound epidermis, distal blastema, and cells at proximal and lateral regions of the blastema. The expressions of msxe (B), ilf2 (D), smarca4 (E), and hspa9 (J) were seen exclusively in the blastema, whereas junbl was only in the distal part of blastema (C). Similarly, the wound epidermis can be subdivided into several distinct regions: the lateral part of basal wound epidermis expressing phlda2 (F), the basal wound epidermis expressing mvp (G), a layer of regenerating epidermis expressing fn1b (H), and the apical epidermis expressing junb (K). Furthermore, mmp9 expression was seen in cells irregularly distributed within the basal wound epidermis and in cells at the proximal and lateral region of the blastema, which appear to overlap with those expressing dlx5a. As in the larval fin fold, the expression of all genes was not detected in uncut fins. Arrowheads mark the sites of amputation. The same magnifications for respective right and left columns of panels A–K (scale bars in panel A). EXPRESSION / LABELING:
|
|
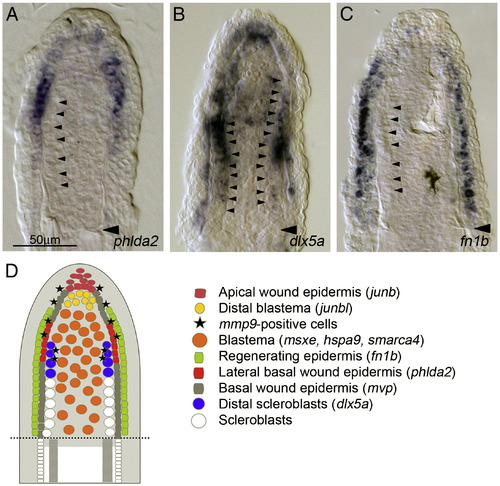
Cellular organization of regeneration niche in adult regenerates. (A–C) Large magnification of fin sections in which the expressions of phlda2 (A), dlx5a (B) and fn1b (C) were detected. phdla2 expression is seen at the lateral part of basal wound epidermis, with a gradient of weak expression to distal and proximal directions (A); whereas dlx5a expression shows a strong expression in cells proximal–lateral part of blastema in addition to a uniform weak expression in the basal wound epidermis. Judging from the continuous alignment of the large-sized cells (arrowheads) to the distal side of fin rays and from the coincidence with expressions of runx2a, runx2b, bmp6, bmp2b and ihha (Smith et al., 2006), these cells with strong dlx5a expression appear to be the early scleroblasts beginning to differentiate into the fin rays. The same magnifications for panels A–C (scale bar in panel A). (D) A schematic drawing showing the cellular organization in the adult fin regeneration niche. From the in situ expression analysis of regeneration-induced genes, at least 8 distinct cell types, 4 cell types in the wound epidermis, 3 in the blastema, and 1 unknown mmp9-positive cells, appear to be present during regeneration. EXPRESSION / LABELING:
|
|
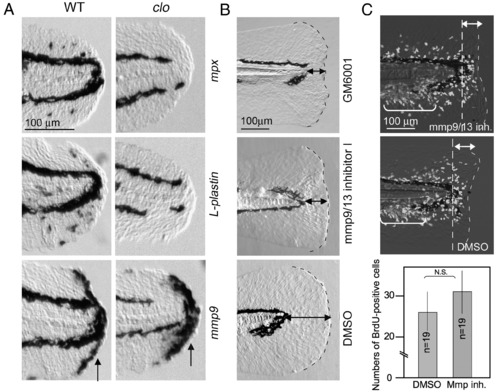
Role of Mmp9 expressed by non-myeloid cells during regeneration. (A) Normal expression of mmp9 in clo mutants, in which the myeloid cell differentiation is impaired. The expressions of mpx (top row), L-plastin (middle row) and mmp9 (bottom row) were detected in wild type larvae and clo mutants at 6 hpa. No significant decrease was observed for mmp9 expression (arrows), whereas the expression of mpx and L-plastin was undetectable (n = 8, respectively). The gene expression was confirmed by using 7–9 mutant larvae for the respective probes. (B) Abnormal larval regeneration caused by inhibitors of Mmps. Larvae exposed to GM6001 (0.1 mM, top) and Mmp9/13 inhibitor I (0.1 mM, middle) showed a deformed and irregular regeneration (9/9 for GM6001, and 19/32 for Mmp9/13 inhibitor I), whereas the amputated larvae exposed to the vehicle did not display any defect (bottom). Similar inhibitory effect was also observed using Mmp9-specifc inhibitor I (0.1 mM, 10/13, data not shown). The same magnifications for respective panels in A or B (scale bars in the top row). (C) Normal activation of cell proliferation in the presence of Mmp9 inhibitor. BrdU labeling was carried out 18–24 hpa in the presence (top) and absence (middle) of Mmp9 inhibitor. Significant difference of cell proliferation was not detected. Many of responding cells to amputation are present in areas indicated by double-headed arrows. The bracketed areas represent cells that fate to form the adult tail fins. The same magnifications for top and middle panels (scale bar in the top). Quantification (bottom) was carried out by counting BrdU-positive cells in the caudal fin fold (posterior to the dashed lines). Data in the graph are the mean ± SEM. Statistical significance was tested by Student's t-test at P = 0.01. N.S., not significant. EXPRESSION / LABELING:
|
|
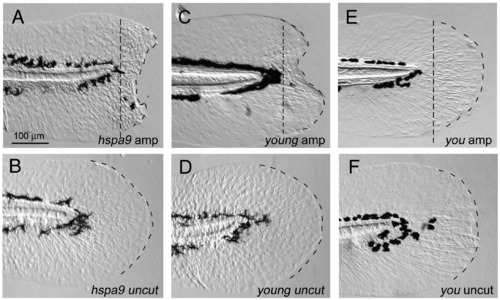
Genetic mutations affecting the larval fin fold regeneration. (A, B) Disrupted larval fin fold regeneration in hspa9 mutants. The larval regeneration was apparently impaired (severe defect in 44/56 mutants, mild defect in 8/56 mutants; (A), whereas the uncut mutants did not show such defects (B). (C, D) Disrupted larval fin fold regeneration in yng/smarca4 mutants. A complete penetrance of defect in regeneration was observed in yng/smarca4 mutants (n = 28/28 mutants, C). Despite the mutant phenotypes such as smaller eyes and curved body axis, hspa9 and yng/smarca4 mutants were alive at 5 dpf and retain a good cellular viability. (E, F) Normal regeneration in other mutants. Several other mutants including you mutants, which have a defect in hedgehog signaling (Kawakami et al., 2005), did not display such defects (n = 15, E). The same magnifications for panels A–F (scale bar in panel A). PHENOTYPE:
|
|
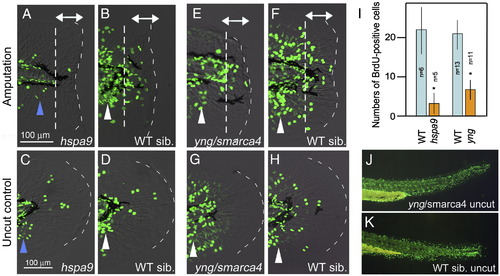
Functions of Hsp9 and Smarca4 are required for the blastema cell proliferation. (A–D) BrdU-incorporation in hspa9 mutants (A, C) and wild type sibling (B, D). Regeneration-dependent cell proliferation in response to amputation (double headed arrow in panel B) was apparently decreased in hspa9 mutants (A). Note that the regeneration-independent cell proliferation, in particular the cell proliferation at the caudal and ventral area, which gives rise to adult-type caudal fin (arrowheads in panels A–D), was also affected in this mutant (A, C). (E–H) BrdU-incorporation in yng/smarca4 mutants (E, F) and wild type sibling (G, H). Regeneration-dependent cell proliferation (double-headed arrows in panel F) was also apparently decreased in this mutant (E), but the regeneration-independent normal growth was only slightly affected (arrowheads in panels E–H). The same magnifications for panels A, B, E, F (scale bar in panel A) and panels C, D, G, H (scale bar in panel C). (I) Quantification of cell proliferation. Counting of BrdU-incorporated cells was carried out only in caudal fin fold (indicated by double-headed arrows) to exclude the regeneration-independent cell proliferation. Data in the graph are the mean ± SEM. Statistical significance was tested by Student's t-test. *P < 0.01. (J, K) BrdU incorporation in the trunk regions of yng/smarca4 mutants (J) and wild type sibling (K). BrdU incorporation in yng/smarca4 mutants was indistinguishable from that of wild type siblings in other body regions, suggesting that the mutants have no apparent defect in normal cell proliferation. |
|
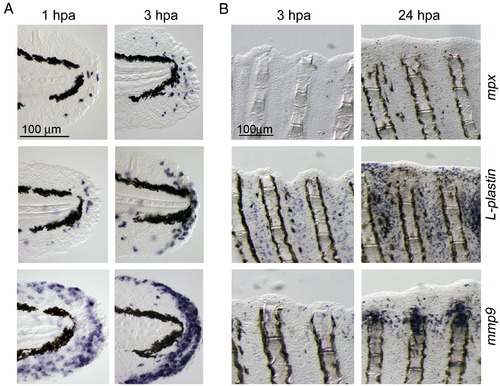
Distribution of neutrophils, macrophages, and mmp9-positive cells during larval and adult regeneration. (A) ISH analyses for expression of mpx, a neutrophils marker (top row), L-plastin, a macrophage marker (middle row) and mmp9 (bottom row) during larval fin fold regeneration. (B) Expression of mpx, L-plastin and mmp9 during adult fin regeneration. Only a few mpx-positive neutrophils were seen during regeneration up to 24 hpa (top row), whereas a large number of macrophages (middle row) were recruited to regions around the wound extending over 1 ray segment away from the stump. The mmp9-positive cells appeared by 24 hpa, however many mmp9-positive cells were localized at the amputation site and adjacent inter-ray regions with apparently different expression profile from mpx or L-plastin. The same magnifications for respective panels in A and B (scale bars in top left panels). |
|
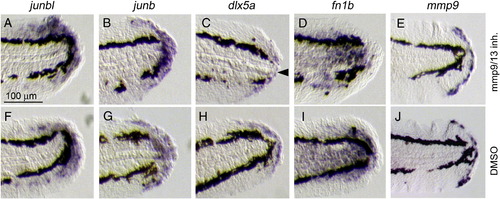
Formation of the blastema, wound epithelium and other regeneration-induced cell types in the presence of Mmp inhibitors. (A–J) ISH analysis for the induction of the respective genes after exposure to Mmp9/13 inhibitor I (A–E) or to the vehicle (F–J) at 1 dpa. The expressions of junbl (A, F), junb (B, G), dlx5a (C, H), fn1b (D, I) and mmp9 (E, J) were not abolished by the inhibitor, suggesting that the formation of respective cell groups in response to amputation was not impaired by the Mmp inhibitors. Note that a slight decrease of dlx5 expression (arrowhead), implying that the inhibitor could have an effect on the proper formation of wound epithelium. The gene expression was confirmed by using 7–10 drug-treated larvae for the respective probes. The same magnifications for all panels (scale bar in A). |
|
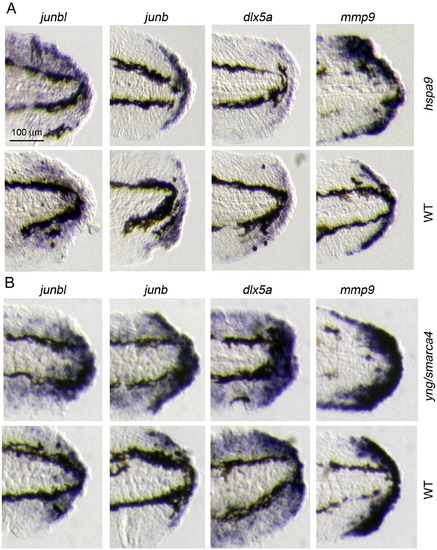
ISH analysis of hspa9 and yng/smarca4 mutants. Respective gene expressions were detected in mutants (upper panels) and wild type siblings (lower panels). 5–7 mutant larvae were used for each probe. |
|
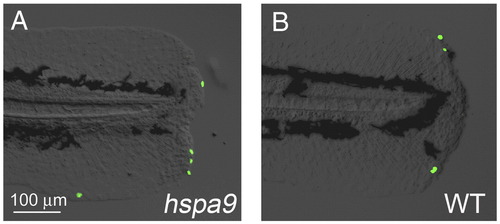
Cellular apoptosis was not affected during larval fin fold regeneration in hspa9 mutants. (A, B) Apoptosis was assessed by TUNEL staining in hspa9 mutants (A) and wild type larvae (B) at 1 dpa. No increased apoptosis was observed. |

Unillustrated author statements PHENOTYPE:
|
Reprinted from Developmental Biology, 325(1), Yoshinari, N., Ishida, T., Kudo, A., and Kawakami, A., Gene expression and functional analysis of zebrafish larval fin fold regeneration, 71-81, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.