- Title
-
Impaired mitophagy links mitochondrial disease to epithelial stress in methylmalonyl-CoA mutase deficiency
- Authors
- Luciani, A., Schumann, A., Berquez, M., Chen, Z., Nieri, D., Failli, M., Debaix, H., Festa, B.P., Tokonami, N., Raimondi, A., Cremonesi, A., Carrella, D., Forny, P., Kölker, S., Diomedi Camassei, F., Diaz, F., Moraes, C.T., Di Bernardo, D., Baumgartner, M.R., Devuyst, O.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|
|
PHENOTYPE:
|
|
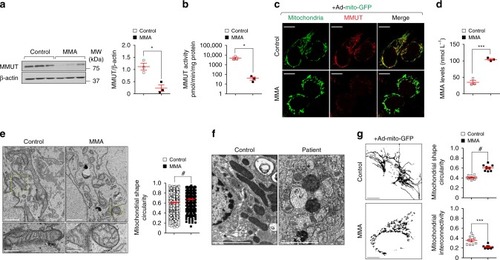
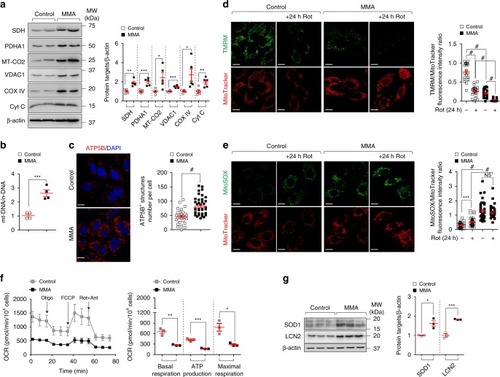
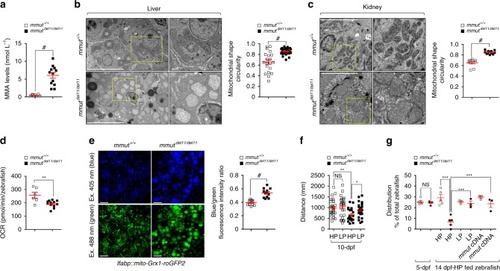
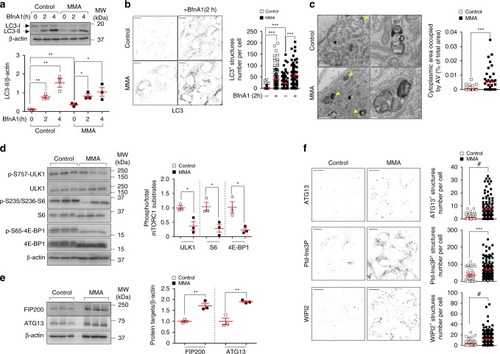
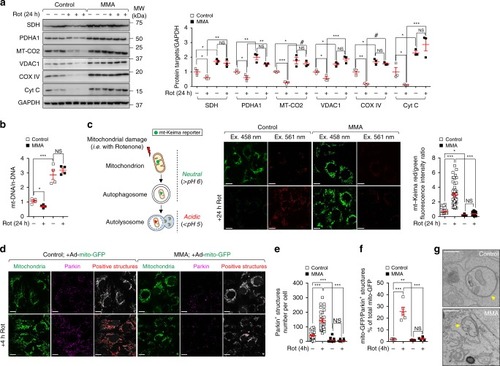
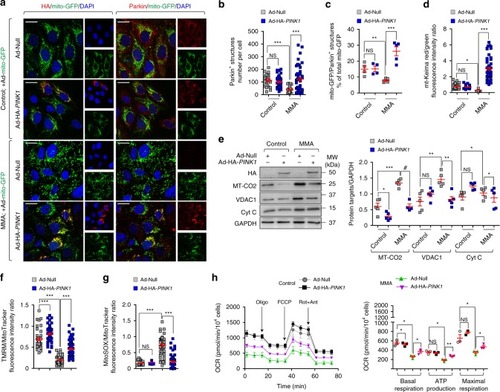
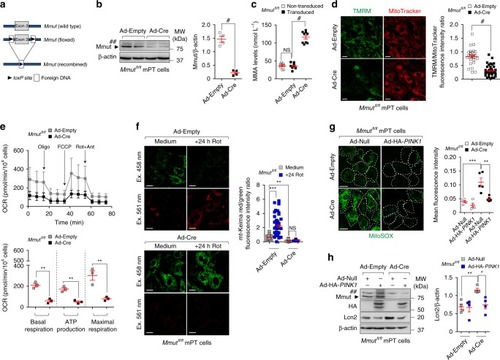
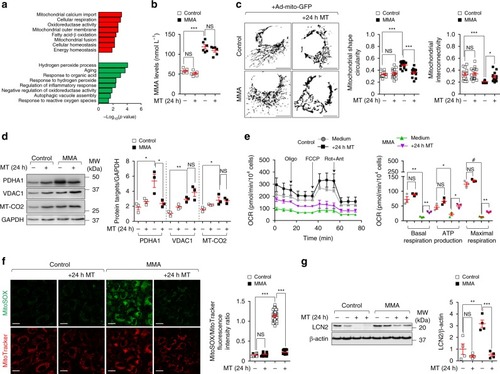
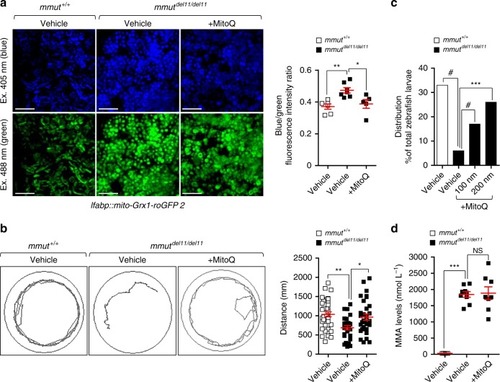
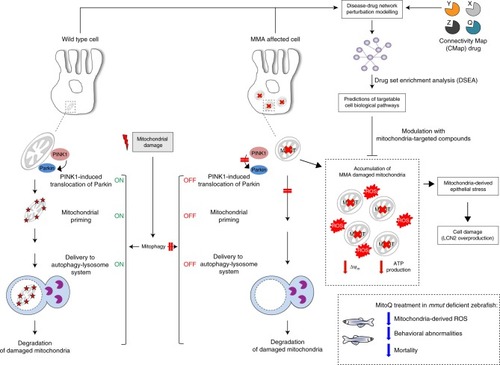
In wild-type kidney cells (left), mitochondrial stress (e.g. treatment with Rotenone) stimulates PINK1-induced translocation of Parkin to damaged mitochondria. This triggers mitophagy and the subsequent disposal of dysfunctional mitochondria through autophagy–lysosome degradation systems, thereby safeguarding the homeostasis and function of the mitochondrial network. By contrast, in MMA-affected kidney cells (right), the impaired PINK/Parkin-mediated mitophagy impedes the delivery of damaged mitochondria and their degradation by autophagy‒lysosome pathways. This leads to accumulation of MMA-diseased mitochondria and exacerbates the mitochondrial alterations induced by |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |