- Title
-
Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon
- Authors
- Zambusi, A., Pelin Burhan, Ö., Di Giaimo, R., Schmid, B., Ninkovic, J.
- Source
- Full text @ Cells
|
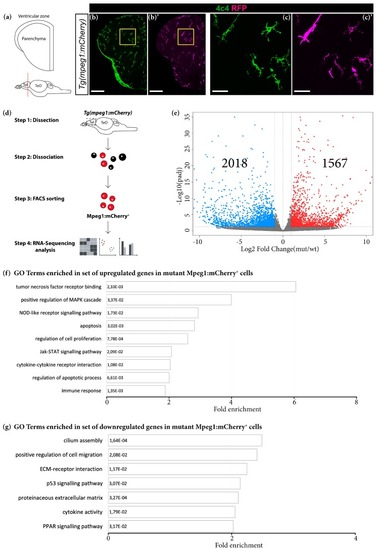
Grna and Grnb deficiency leads to transcriptional changes associated with an activated microglial state. ( |
|
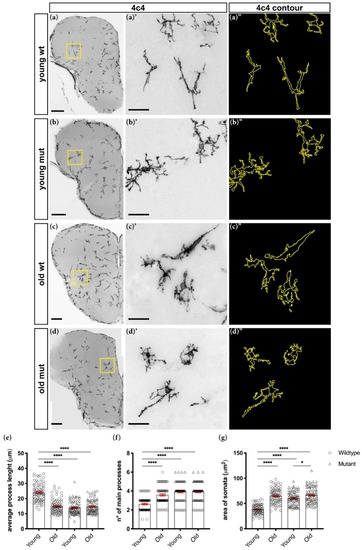
Grna; Grnb-deficient microglial cells display morphological changes associated with aging. ( PHENOTYPE:
|
|
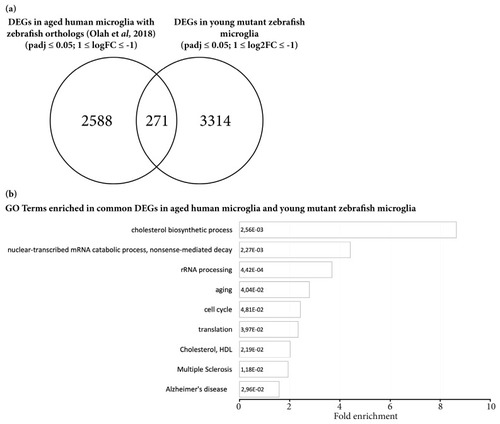
Microglial cells isolated from young mutant zebrafish share a proportion of DEGs with aging human microglial cells. ( |
|
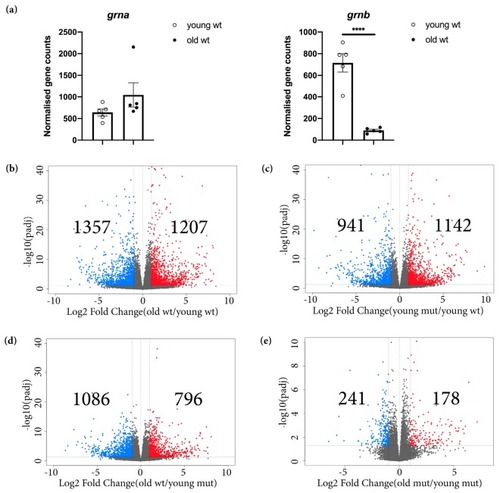
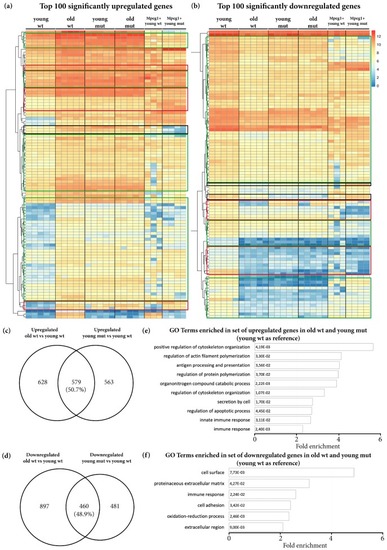
Transcriptomic changes detected in old wildtype, young mutant, and old mutant telencephali. ( EXPRESSION / LABELING:
|
|
The phenotype of age-related transcriptional changes detected in old wildtype animals is partially mimicked by Grna and Grnb deficiency in young mutant animals. ( |
|
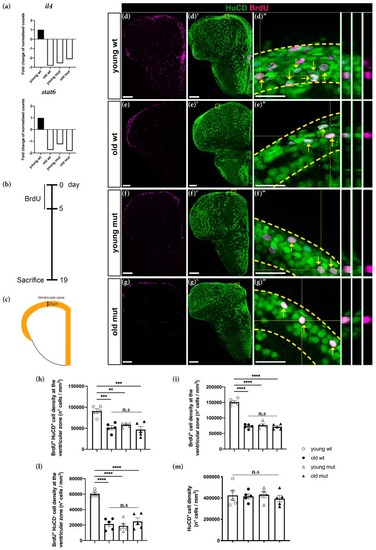
Grna and Grnb deficiency mimics the phenotype of the decrease in neurogenesis observed during aging in the adult zebrafish telencephalon. ( |
|
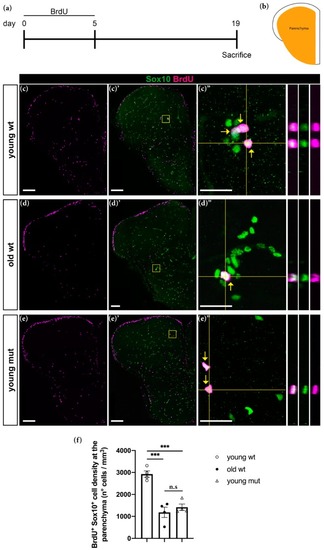
Grna; Grnb deficiency mimics the phenotype of the decrease in oligodendrogenesis observed during aging in the adult zebrafish telencephalon. ( PHENOTYPE:
|
|
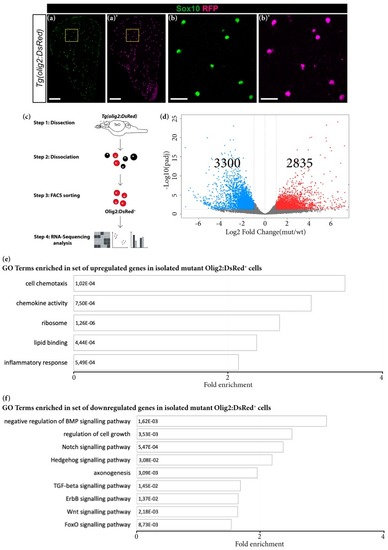
Transcriptomic changes in FACS-isolated mutant oligodendroglial cells. ( |
|
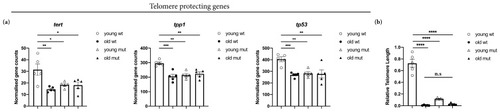
Grna and Grnb deficiency causes downregulation of telomere-protective genes and telomere shortening in the telencephalon in young zebrafish, as observed in old animals. ( |