- Title
-
Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent
- Authors
- Patra, C., van Amerongen, M.J., Ghosh, S., Ricciardi, F., Sajjad, A., Novoyatleva, T., Mogha, A., Monk, K.R., Mühlfeld, C., and Engel, F.B.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
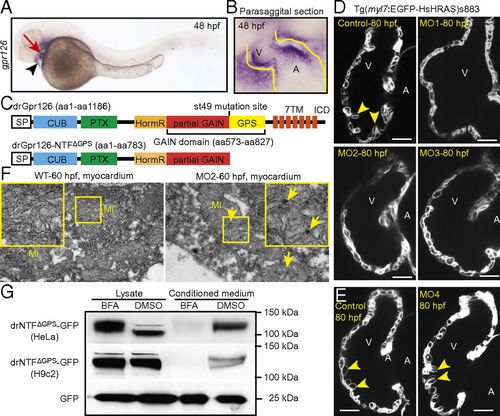
Gpr126 knockdown leads to cardiac abnormalities in zebrafish. (A) Lateral view of a 48-hpf zebrafish embryo after whole-mount in situ hybridization showing gpr126 expression in heart (red arrow) and pericardium (black arrowhead). (B) Parasagittal section confirming cardiac expression of gpr126. (C) Schematic representation of zebrafish full-length Gpr126 (drGpr126) and its NTF part up to the st49 mutation site (drGpr126-NTFΔGPS). (D and E) Confocal sections of hearts from control- and morpholino-injected Tg(myl7:EGFP-HsHRAS)s883 embryos at 80 hpf. In full-length Gpr126-depleted animals (MO1-3) but not in Gpr126-CTF–depleted (MO4) morphants, trabeculation (yellow arrowheads) is perturbed. (Scale bar: 20 μm.) (F) Transmission electron micrographs at 60 hpf reveal that morphants contain elongated Mi with more branched cristae and electron dense precipitates (arrows) compared with WT siblings. (G) Western blot analysis of lysates and conditioned medium from cells overexpressing C-terminal GFP-tagged drGpr126-NTFΔGPS or GFP. Note that both GFP-tagged NTFΔGPS (predicted band size: 113 kDa) and GFP (predicted band size: 27 kDa) were detected in conditioned medium. However, secretion of GFP-tagged NTFΔGPS but not GFP was inhibited by BFA (a blocker of classical trans-Golgi secretory pathway). A, atrium; V, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
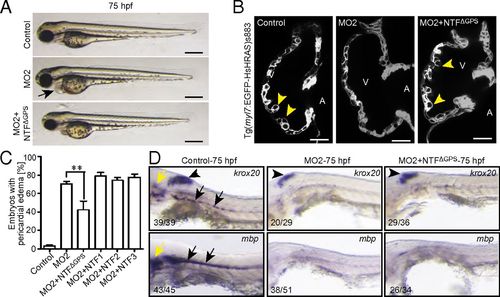
NTFΔGPS mRNA rescues the trabeculation phenotype in Gpr126-depleted zebrafish. (A) Lateral view of control-, MO2-, or MO2+NTFΔGPS mRNA-injected embryos at 75 hpf. MO2 injection resulted in pericardial edema (arrow). (B) Confocal sections of the hearts from control-, MO2-, and MO2+NTFΔGPS mRNA-injected Tg(myl7:EGFP-HsHRAS)s883 embryos at 80 hpf. In Gpr126-depleted animals, trabeculation (yellow arrowheads) is perturbed. Note that this phenotype is rescued by NTFΔGPS mRNA injection. (C) Quantitative analysis of rescue experiments of coinjections of MO2 and mRNAs encoding NTFΔGPS or NTF subfragments. Mean ± SEM. (D) Whole-mount in situ hybridization demonstrating krox20/egr2 and mbp expression in the otic vesicle (yellow arrow) and PLLn Schwann cells (black arrow) and krox20/egr2 expression in the hindbrain (arrowhead) at 75 hpf. In contrast, expression of krox20/egr2 and mbp is down-regulated in PLLn of MO2-mediated full-length Gpr126-depleted embryos. Coinjection of NTFΔGPS mRNA failed to rescue the PLLn myelineation phenotype in the cardiac phenotype-rescued animals. **P < 0.01. EXPRESSION / LABELING:
PHENOTYPE:
|
|
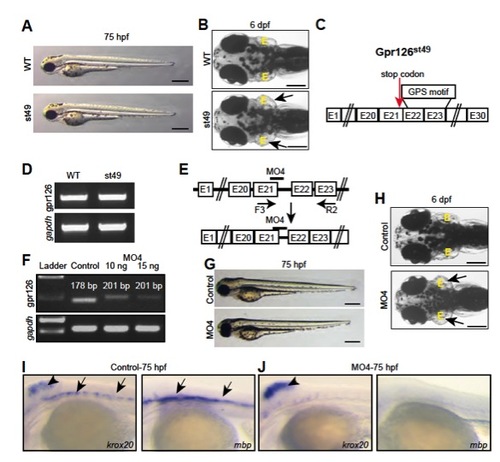
Inhibition of the 7TM domain of zebrafish Gpr126 does not disrupt heart morphogenesis. (A) No pericardial edema is observed at 75 h postfertilization (hpf) in gpr126st49 zebrafish mutant embryos. (Scale bar: 400 μm.) (B) Dorsal view of a WT and a gpr126st49 mutant larva at 6 d postfertilization (dpf). gpr126st49 mutant larva showing enlarged ears (arrows). (Scale bar: 200 μm.) (C) Schematic depiction of zebrafish gpr126st49 mutant mRNA. The mutation introduces a premature stop codon before the GPS motif (red arrow). (D) RT-PCR analysis showing that gpr126 mRNA transcripts could be detected in gpr126st49 mutants. gapdh was used as loading control. (E) Schematic depiction of zebrafish prespliced gpr126 mRNA. Exons are represented by boxes, and introns are represented by lines. Bar represents the MO4 binding site. Arrows below the respective Exons indicate primer binding sites; these are used to analyze the fate of prespliced gpr126 mRNA after MO4 injection. (F) RT-PCR analysis of mRNAs from WT and MO4-injected 3-dpf zebrafish embryos using primers as indicated in E. gapdh was used as loading control. MO4 injection resulted in partial intron insertion. (G) MO4 injection did not cause pericardial edema at 75 hpf. (Scale bar: 400 μm.) (H) Dorsal view of a phenol red-injected WT larva and a MO4-injected larva at 6 dpf. MO4-injected larva showing enlarged ears (arrows). (Scale bar: 200 μm.) (I and J) Whole-mount in situ hybridization demonstrating krox20/egr2 [hindbrain, arrowhead; posterior lateral line nerve (PLLn) Schwann cells, arrows] and mbp (PLLn Schwann cells, arrows) expression at 75 hpf. MO4 morphants, which express NTFΔGPS but not CTF, exhibit normal expression of krox20/egr2 in the hindbrain (arrowhead), but both krox20/egr2 and mbp expression are down-regulated in the PLLn at 75 hpf. E, ear. EXPRESSION / LABELING:
PHENOTYPE:
|
|
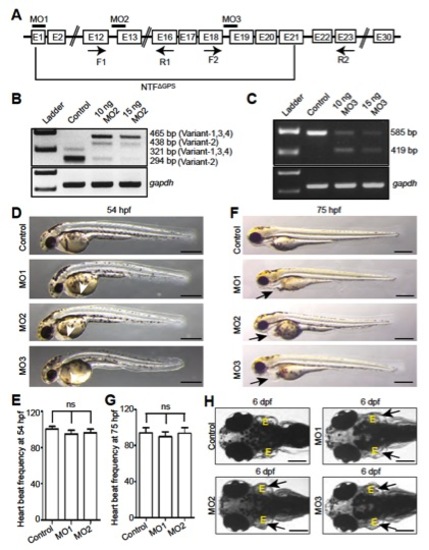
Full-length gpr126 knockdown causes pericardial edema. (A) Schematic depiction of zebrafish prespliced gpr126 mRNA. Exons are represented by boxes, and introns are represented by lines. Bars represent the different morpholino binding sites. Arrows below the respective exons indicate primer binding sites; these are used to analyze the efficiency of gene knockdown by RT-PCR. (B and C) RT-PCR analysis of cDNA made from RNA isolated from 2-dpf control and MO2- or MO3-injected embryos using F1 and R1 primers for MO2 (B) and F2 and R2 primer for MO3 (C) as indicated in A. gapdh was used as loading control. MO2 binding resulted in 144-bp-long intron insertions (B). MO3 binding caused 166-bp-long exon deletions (C) causing a frame shift. (D and F) Brightfield images of control- or morpholino-injected embryos at 54 hpf (D) and 75 hpf (F). gpr126 knockdown resulted in mild accumulation of blood at the sinus venosus (arrowheads) at 54 hpf (D). Prominent pericardial edema (arrows) was observed at 75 hpf in Gpr126-depleted embryos (F). (Scale bar: 400 μm.) (E and G) Quantification of cardiac contraction frequency of phenol red (control)- or morpholino-injected embryos at 54 hpf (E) or 75 hpf (G). Cardiac contraction frequency of the phenol red-injected embryos was set to 100%. (H) Dorsal view of a phenol red-injected WT larva and MO1-, MO2-, or MO3-injected larva at 6 dpf. MO1, MO2, and MO3 injection all caused enlarged ears (arrows). (Scale bar: 200 μm.) ns, not significant; NTFΔGPS, N-terminal fragment up to the st49 mutation site. |
|
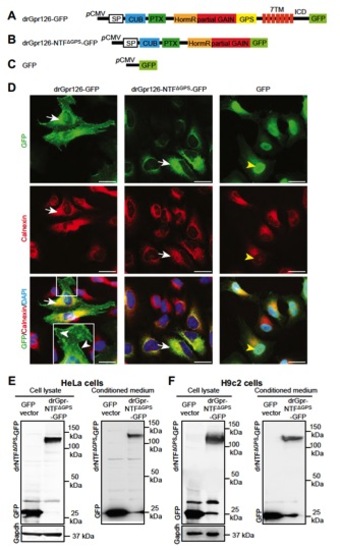
The NTF part of the st49 mutant form of zebrafish Gpr126 is an independent, stable, secreted protein. (A and B) Schematic depiction of the C-terminal GFP-tagged drGpr126 (A) and drGpr126-NTFΔGPS (B). ICD, intracellular domain. (C) GFP vector used as control. (D) Immunofluorescence analysis of HeLa cells stained for GFP (green) and calnexin (red) after transfection with constructs described in A–C. Nuclei were stained with DAPI (blue). drGpr126-GFP localized to the plasma membrane (white arrowheads) and colocalized with calnexin (arrows), an endoplasmic reticulum marker protein. drGpr126-NTFΔGPS-GFP was not detected at the plasma membrane but colocalized with calnexin (arrows). GFP was predominantly expressed in the nucleus (yellow arrowheads). (E and F) Western blot analysis of cell lysates and conditioned medium from HeLa (E) and H9c2 cells (F) after transfection with C-terminal GFP-tagged drGpr126-NTFΔGPS or GFP. Note that GFP-tagged NTFΔGPS (predicted band size: 113 kDa) and GFP (predicted band size: 27 kDa) were detected in both lysate and conditioned medium. Gapdh was used as loading control for cell lysates. |