- Title
-
The chromosomal passenger protein birc5b organizes microfilaments and germ plasm in the zebrafish embryo
- Authors
- Nair, S., Marlow, F., Abrams, E., Kapp, L., Mullins, M.C., and Pelegri, F.
- Source
- Full text @ PLoS Genet.
|
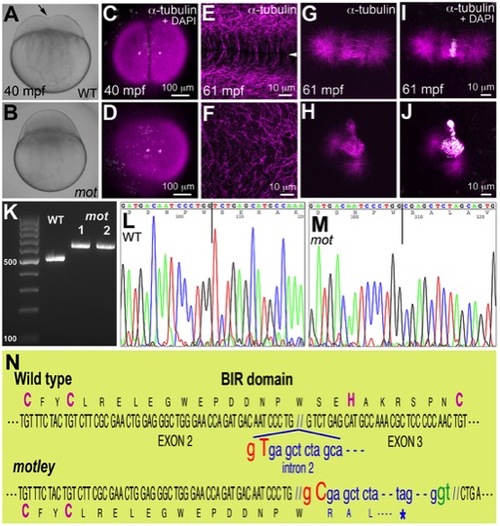
motley is a mutation in birc5b, which causes early cell division defects in zebrafish embryos. The cleavage furrow (arrow) seen in live wild-type embryos at 40 mpf during the first cell cycle (A) is absent in motley mutants (B). Early embryonic cytokinesis in zebrafish are rapid and overlap with each other with furrow completion of the previous cell cycle occurring concurrently with the furrow initiation of the next. This results in furrows of various stages of development being present at a single time point in the same embryo (e.g. Figure S1E). During the early cell cycles, immunolabeling for α-tubulin reveal abutting microtubule arrays between nuclei in wild-type blastodisc (C, shown for the furrow corresponding to the first cell cycle), which are abnormal in motley (D). DAPI channel in (C,D) reveals nuclear cycle progresses normally in mutants, although separation of daughter nuclei is reduced likely due to spindle defects (scattered white speckles are caused by autofluorescence by yolk granules, which typically can not be fully removed in the mounting preparation). High magnifications shown here corresponding to the second cell cycles in wild-type and motley show the microtubule-free zone at the furrow in wild-type blastodisc (arrowhead in E, low magnification in Figure S1A), which does not exist in motley (F, low magnification in Figure S1C, boxed area rotated 90° in F). Wild-type mitotic spindle aligns the DNA at the center of the spindle (G, I) while spindles in motley mutants are bent with the DNA spread along their length (H, J). RT-PCRs show that wild-type birc5b is ~500 bp, while motley allele from two different homozygous mutant females is larger at ~600 bp (K). Sequence chromatograms of wild-type (L) and motley (M) birc5b alleles show that the motley sequence is unaltered up to position 236 from the start ATG (vertical line, corresponding to base 305 in the shown sequence chromatograms) and thereafter corresponds to intronic sequence. Schematic of the Birc5b BIR domain that is disrupted due to intron insertion in motley (N). |
|
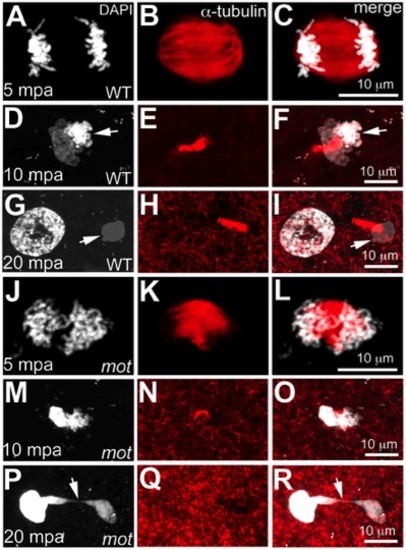
motley mutant eggs fail to complete meiosis II. In water-activated wild-type eggs, sister chromatids align at meiotic spindle poles during anaphase (A–C). The meiotic spindle bundles into a nascent midbody between one set of condensing (D, F arrow) and one set of decondensing chromatid sets (D–F). The smaller, condensed polar body (G, I arrow) and the decondensed, larger female pronucleus separate with the meiotic midbody attached to the polar body (G–I). In motley eggs, sister chromatids spread along the bent meiotic spindle (J–L). Sister chromatid sets cannot be distinguished as pronucleus or polar body and a smaller meiotic midbody is occasionally seen in motley (M–O). Sister chromatids attempt to separate but remain connected by chromosomal bridges spanning several microns (P–R, arrows). PHENOTYPE:
|
|
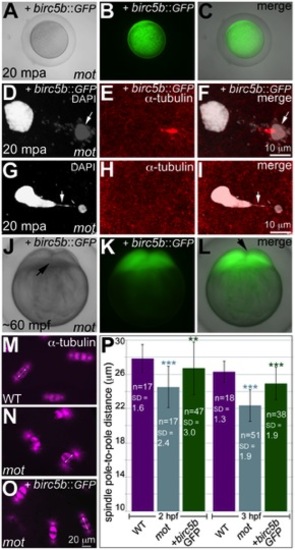
Wild-type birc5b rescues cell division defects in motley mutants. In vitro cultured oocytes from homozygous motley females express the injected birc5b::eGFP, mature and water-activate normally (A–C). Birc5b::eGFP-expressing motley eggs successfully complete meiosis as seen by the presence of meiotic midbody (E, F) and a distinctive polar body (D, F arrow). Control uninjected motley eggs do not have a meiotic midbody and form chromosomal bridges (G, I arrow) between condensed DNA masses (G–I). Normal cleavage furrows at ~60 mpf in in vitro fertilized embryos from Birc5b::eGFP-expressing eggs (J–L, arrows). Normal mitotic spindles in wild-type embryos (M), bent spindles in motley mutants (N), and mitotic spindles in Birc5b::eGFP-expressing motley embryos resembling wild-type (O). Due to the bending of the spindle, linear spindle pole-to-pole distance (SPD) is shorter in motley mutants than wild-type, while in Birc5b::eGFP-expressing motley embryos this distance approaches wild-type (P). Double-headed arrows in M-O indicate example SPD distances used for quantitation in P. Error bars in P represent Standard Deviation. Statistical significance of SPD values was ascertained by t-test (2-tailed, unpaired, unequal variance). P<0.001 = ***, P<0.01 = **. SPD distances were compared between uninjected motley and wild-type at 2 and 3 hpf (light blue asterisks), and between uninjected motley and birc5b::GFP injected motley at 2 and 3 hpf (green asterisks). P values were not significant (P>0.05), between wild-type and rescued birc5b::GFP injected SPDs. |
|
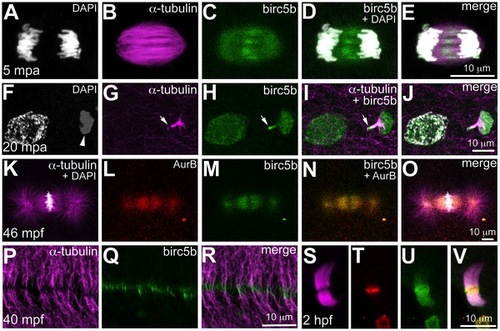
Birc5b protein is expressed during meiosis, mitosis, and cytokinesis. Birc5b is expressed along the length of the meiotic spindle during anaphase and concentrates at the spindle midzone (A–E). Birc5b localizes to the meiotic midbody and concentrates at one end, corresponding to the ooplasmic side (F–J, arrow in H, I). During mitotic metaphase Birc5b co-localizes with AurB on the spindle (K–O). During cytokinesis in the early embryo, Birc5b localizes to the tips of bundled microtubules at the furrow (P–R). At the midbody in the cleavage-stage embryo, shown here at 2 hpf (S–V), Birc5b expression (U) overlaps with AurB (T) at the center of bundled microtubules (S) and extends beyond it. EXPRESSION / LABELING:
|
|
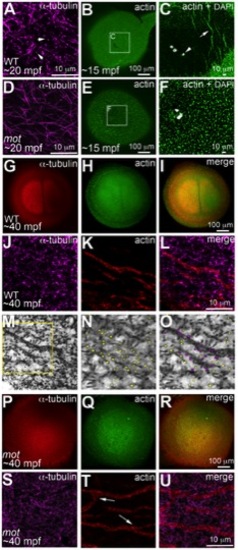
motley/birc5b mutants fail to rearrange cortical actin microfilaments. Animal views of blastodisc cortex. In wild-type embryos (A–C) tips of the sperm monoaster microtubules are found at the cortex (A, arrows). Microtubule punctae are seen at the cortex, which are the ends of astral microtubules present in focal planes beneath the cortex (not shown). Short F-actin seed filaments polymerize (C, arrow) and clear from the center of the wild-type blastodisc (B, C; polar body, arrowhead in C), and congressing pronuclei are also apparent). In motley/birc5b mutants, sperm monoaster microtubules tips cannot be detected at the cortex (D), and polymerizing F-actin seed filaments fail to clear from the center (E, F). At 40 mpf in wild-type embryos, astral microtubules of the first mitotic spindle radiate towards the blastodisc cortical periphery (G) while the microfilaments are seen at the cortical periphery in concentric rings (H, I). The tips of the mitotic aster microtubules are also seen at the cortex at this time, (J) where they contact peripheral microfilaments (K, L). Panel M is a color-inverted black and white image of J, a region of which has been expanded in (N,O). In (N), microtubule tips in J and M are indicated by yellow dots, while in (O) microtubule tips from N that additionally appear in contact with the microfilaments have been colored magenta. Mitotic astral microtubules fail to separate in motley (P) and the microfilaments are found aberrantly in the center of the cortex and are branched (Q, R). The tips of motley mitotic aster microtubules are also absent at the cortex (S), while microfilaments in motley are aberrantly branched (T, U, arrows in T indicate ectopic branching) and do not appear to colocalize with microtubules (U). Panels C and F are high magnifications of indicated boxes in B, E, respectively. Panels A, D, (J–O) and (S–T) are high magnifications of the cortex from blastodiscs of stages comparable to those in B, E, I and R, respectively. Reduced labeling intensity of the right aster in G is an imaging artefact due to slight tilting of the blastodisc within the semi-flat mount preparation. PHENOTYPE:
|
|
Germ plasm RNPs localize to the tips of astral microtubules. Animal views of blastodisc cortex. (A–C) GP RNPs are located in a band (B) at the cortical periphery. Higher magnification images indicate that multimerized GP RNPs are located at the tips of spindle pole astral microtubules (D–F; G–I). In non-furrow peripheral regions (D–F), several microtubule tips (D) can be observed to coincide with a large multimerized GP RNP (E, F). Arrow in F points to a smaller multimerized aggregate at a microtubule tip. At the cleavage furrow (G–I), multimerized GP RNPs are found as bilateral rows of aggregates (arrows in I) at bundled tips of astral microtubules from contralateral sides (G–I). Panels G–I are high magnification views of the rectangular box (labeled I) in panel C, rotated 90° counterclockwise. Panels D–F are high magnification views from an embryo comparable to that in C of a non-furrow region similar to the square box indicated in panel C (labeled F). EXPRESSION / LABELING:
|
|
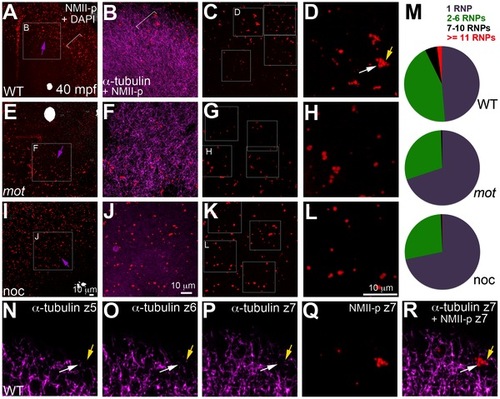
motley/birc5b mutants fail to effectively multimerize GP RNPs. Animal views of blastodisc cortex. In wild-type embryos (A–D) the GP RNP aggregation wave (bracket, A) coincides with cortical astral microtubule ends (B, bracket indicates the wavefront within the wave). GP RNP multimerization in wild-type embryos result in large GP RNP aggregates (C, D). In motley/birc5b mutants (E–H), cortical microtubules are disorganized (F), and GP RNPs fail to multimerize effectively (G, H). In nocodazole-treated embryos (I–L), cortical microtubules are absent (J) and GP RNP multimerization also fails (K, L). Semi-quantitative analysis of GP RNP aggregation showing similar multimerization failures in motley/birc5b and nocodazole-treated embryos compared to wild-type (M). Inset labels in panels A, E, I and C, G, and K indicate ROIs shown at higher magnifications next to the respective panels. Arrows in A, E and I indicate the radial direction, from the center of the blastodisc, along which microtubules in B tend to be oriented in wild-type embryos. (N–R) consecutive confocal sections acquired with a z-step of 0.5 microns: z5 (N, most cortical), z6 (O) and z7 (P, Q, most internal). Yellow arrow shows a single microtubule observed in Z6 (O), Z7 (P) and more internal (not shown) planes but not in z5 (N), which appears associated with an RNP in a multimerized aggregate (Q, R). White arrow indicates a pair of microtubules that appear to converge on a different set of RNPs in the same aggregate (Q,R). The merge panel in R corresponds to microtubules and RNPs for z7 to show the association of microtubule tips with RNP aggregates. |
|
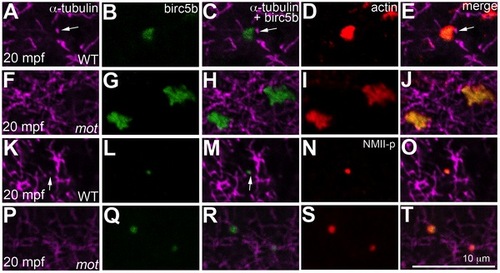
Birc5b localizes to cortical microtubule tips and co-localizes with F-actin and germ plasm RNPs. High magnifications of animal views of blastodisc cortex. In wild-type embryos, Birc5b (B, L) localizes to monoaster microtubule tips (A, C, K, M, arrows) and polymerizing F-actin (D, E). In motley/birc5b mutants (F–J) Birc5b co-localizes with polymerizing F-actin (I, J), but as tips of the microtubules are not seen at the cortex (F), co-localization to the microtubule tip is not observed. Birc5b at the microtubule tips (K, M arrow) co-localizes with GP RNPs in wild-type embryos (N, O). In motley/birc5b mutants (P–T), microtubule tips are not seen at the cortex (P) but Birc5b continues to localize with GP RNPs (S, T). EXPRESSION / LABELING:
|
|
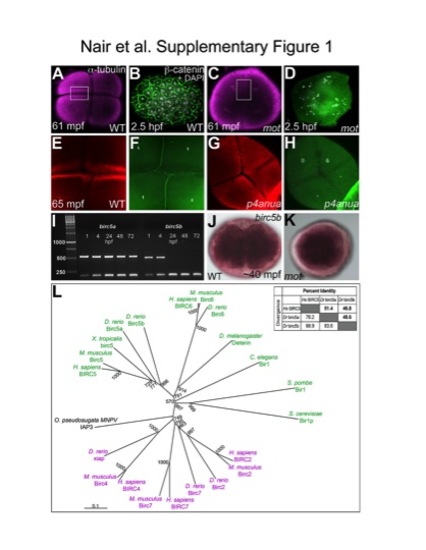
Cell division defects in the maternal-effect mutants motleyp1aiue and p4anua and identification of motleyp1aiue as birc5b. (A–H) Furrow formation defect in motley (A,B,E,F) and p4anua (C,D,G,H) mutants. During the second cell cycle, two intersecting mature cleavage furrows are seen in wild-type embryos (A), and reiterative cytokinesis results in a cellularized blastoderm with distinct nuclei as seen by β-catenin and DAPI labeling (B). During the second cell cycle, the appearance of furrows in motley (C) is due to spreading astral microtubules that never mature into furrows (D). DNA segregation errors in motley manifest as unevenly distributed DNA patches (D). During the second cell cycle, compared to the wild-type embryo (E, F), mature cleavage furrows are absent in p4anua mutants (G, H). (I) birc5a is maternally present and is expressed during all stages of embryonic development, while birc5b transcript is not detectable beyond 24 hpf. β-actin was used as a control transcript which is expressed constitutively during development (200 bp band in all lanes). (J,K) Whole mount in situ hybridizations for birc5b show ubiquitous maternal expression in wild-type (J) and motley embryos (K). (L) TreeView rendering of the ClustalX alignment of BIR proteins from several representative species indicate a trichotomy (black, green and pink groupings) in the BIR family and group zebrafish Birc5a and Birc5b with homologous Birc5 proteins. (A–H, J, K) are animal views of blastodiscs. EXPRESSION / LABELING:
PHENOTYPE:
|
|
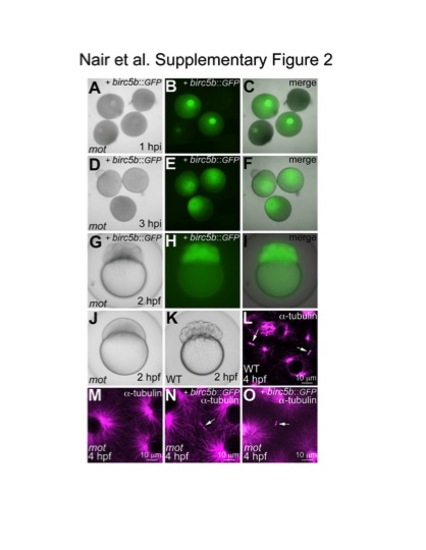
Birc5b::GFP expression in cultured oocytes and late rescue of motley cytokinesis defects by wild-type Birc5b::GFP mRNA injected post-fertilization. (A–F) Expression of Birc5b::GFP through mRNA injection into in vitro matured oocytes. Immature germinal vesicle containing stage IV oocytes from homozygous motley females express protein from injected birc5b::eGFP mRNA 1 hour post injection (hpi) (A–C). Germinal vesicle dissolution and oocyte clearing occur normally in Birc5b::eGFP-expressing motley oocytes (D–F). (G–K) Partial rescue of cytokinesis defects in motley mutants by expression of Birc5b::GFP through mRNA injection into 1-cell embryos. motley mutant embryos injected with birc5b::eGFP at the 1-cell stage are Birc5b::eGFP-positive at 2 hpf and exhibit several cleavage furrows (G–I) like wild-type embryos (K), although blastomeres in mutants are larger due to the lag in functional rescue through injection at the 1-cell stage. Uninjected sibling motley mutants do not exhibit any furrows (J). (L–O) Rescue of midbody formation defect in motley mutants by expression of Birc5b::GFP through mRNA injection into 1-cell embryos. At 4 hpf, midbodies are seen in wild-type embryos (L, arrows), which are never seen in motley mutants (M). In motley mutants injected with birc5b::eGFP at the 1-cell stage, midbodies are seen by 4 hpf (N, O, arrows). Cells in the panels shown (L–O) are at slightly different stages in the cell cycle (shown by different sizes of asters) due to asynchronicity between embryos and embryonic regions characteristic of these stages [53], [54]. However, midbody structures in the early embryo are stable through multiple cell cycles ([21]; our own observations), allowing a comparison between the various conditions. |
|
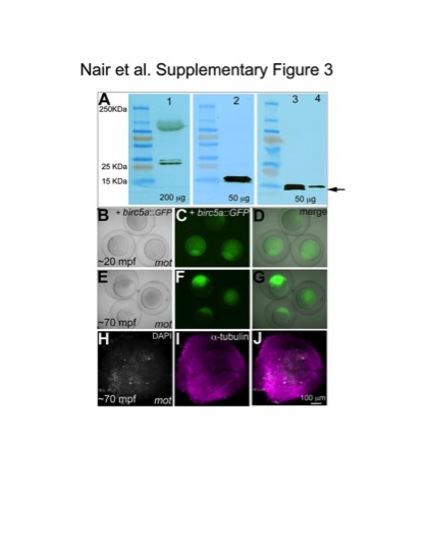
Birc5a and Birc5b protein are both expressed maternally, but are functionally non-redundant. (A) Western blot of whole protein lysates from 20 mpa wild-type eggs (lane 1 and 2), and 40 mpf wild-type (lane 3) and motley embryos (lane 4). Anti-Survivin detects an ~25 KDa protein (lane 1) that is unaffected in motley mutants (data not shown), consistent with this antibody recognizing Birc5a (190 aa, predicted MW 22 kDa). Anti-Survivin-BIR detects a doublet of ~15 KDa protein in wild-type lysates (lanes 2, 3), which is affected in motley mutants (lane 4: lower band missing, upper band with reduced intensity). The mutant product encoded by the mutant motley allele is predicted to include the first 79 aa of the normal protein plus 32 novel aminoacids encoded by intronic sequence (111 aa total, MW ~13 kDa). Because aminoacid composition can affect protein mobility, determination of the precise identity of each band will require protein analysis. However, the data is consistent with anti-Survivin recognizing Birc5a and anti-Survivin-BIR recognizing Birc5b, and a lack of cross-reactivity between these two antibodies and their products. Total protein from each lysate loaded in each lane is indicated in μg. (B–J) Expression of Birc5a::GFP does not rescue the motley/birc5b cytokinesis phenotype. Stage IV oocytes from motley/birc5b mutant females were injected with birc5a::GFP mRNA and in vitro matured into eggs under the same conditions that allowed rescue through expression of birc5b::GFP mRNA. Injected oocytes expressed Birc5a::GFP at ~1hpi and matured into eggs (data not shown). Embryos derived from such Birc5a::GFP-expressing eggs activated normally upon contact with water following in vitro fertilization (B–D) but did not undergo cytokinesis (E–G). Labeling for α-tubulin and DAPI at a time equivalent to the 4-cell stage confirmed that such Birc5a::GFP-expressing, non-cleaving motley/birc5b embryos were indeed fertilized (H–J, note four nuclei showing normal karyokinesis). |
|
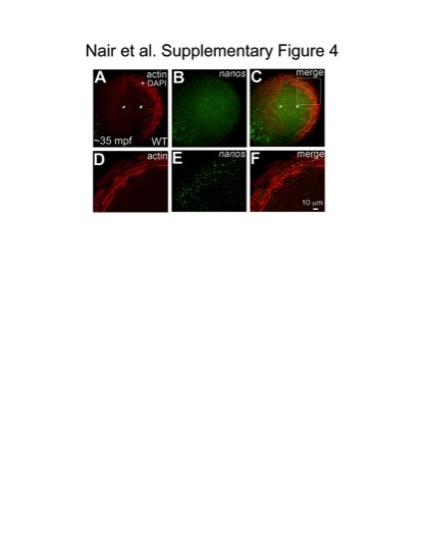
Germ plasm RNPs localize onto cortical microfilaments at the blastodisc periphery. Animal views of blastodiscs; D–F are higher magnifications of area indicated in C, rotated 90° counterclockwise. Fluorescent in situ hybridization for the germ plasm mRNA nanos (B, E), together with immunolabeling for f-actin (A, D) shows that RNPs labeled with nanos co-localize with microfilaments arranged in concentric rings at the cortical periphery (C, F). |
|
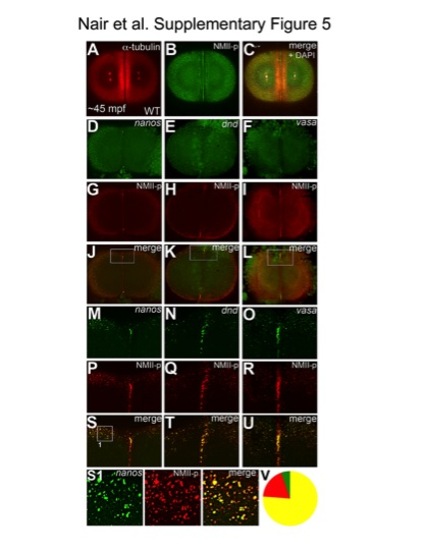
Germ plasm mRNPs are labeled by anti-human phosphorylated non-muscle myosin II antibody. Animal views of 2-cell stage embryos; immunolabelings (A–C), fluorescent in situ hybridizations for nanos (D, M), dnd (E, N) and vasa (F, O), combined with immunolabeling for NMII-p (G–I, P–R). (M–U) Higher magnifications of boxed areas in (J–L). NMII-p labeling recapitulates the known patterns of germ plasm mRNA localization to the distal furrows and peripheral cortex in wild-type embryos at ~45mpf (A–C). nanos, dnd and vasa mRNAs label germ plasm aggregates at the cortical periphery and at the distal ends of the cleavage furrow (D–F, M–O). NMII-p expression overlaps with nanos (J, S), dnd (K, T) and vasa (L, U) both at the outlying cortex and the distal cleavage furrow. (S1) Higher magnification view of an example panel used for the quantitation of colocalization shown in (V), from the image in (S). (V) Pooled counts of observed particles (singletons and multimerized, n = 349) indicate that 76% of particles show both NMII-p and GP RNA labeling (yellow), 19% exhibit only NMII-p labeling (red) and 5% only GP RNA labeling (green). Due to the significantly higher sensitivity of the anti-NMII-p immunofluorescence labeling in comparison to the FISH technique to detect GP RNAs, it is likely that a significant fraction, if not most, of the 19% of particles that exhibit only NMII-p labeling also contain undetected GP RNAs. While we can not rule out that at these stages a minority of particles contain NMII-p without GP RNAs, the observation that less than 5% of particles apparently containing GP RNAs are not labeled with the anti-NMII-p antibody indicates that NMII-p is a reliable marker for GP RNPs. |
|
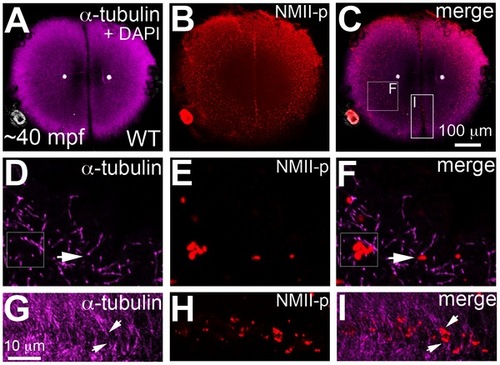
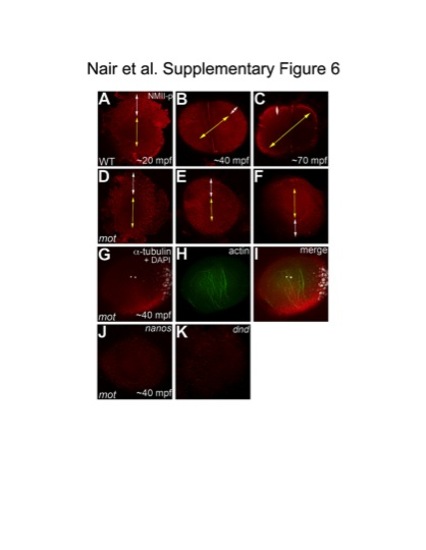
The cortical band of germ plasm RNPs fails to undergo a peripherally-directed compression in motley/birc5b mutants. Animal views of blastodisc cortex, immunolabeled with anti-NMII-p antibody. (A–F) Segregation of GP RNPs during the first two cycles. In wild-type embryos at 20mpf, the center of the blastodisc cortex is GP RNP-free as the RNPs are located in a broad peripheral band (A). As development proceeds, the blastodisc center remains free of GP RNPs and the peripheral band becomes compressed from the center outwards as the central GP RNP-free zone expands (B, C). In motley/birc5b mutants at 20mpf, the center of the blastodisc is GP RNP free and the RNPs are in a broad peripheral band as in wild-type embryos (D). However, the peripheral GP RNP band fails to further compress noticeably during development (E, F). Yellow double-headed arrows represent GP RNP-free zone in the blastodisc center, which does not expand in motley mutants. White double-headed arrows represent peripheral compression of GP RNP band in wild-type embryos, which does not occur effectively in motley/birc5b. (G–I) In motley mutants at a stage coincident with observed GP RNP segregation defects, F-actin (H,I) forms large bundles randomly crisscrossing the blastodisc, instead of circumferential bundles as observed in wild-type (Figure 5B, 5C, 5H; [16]). Such randomly oriented bundles also form in embryos after inhibition of microtubule polymerization [16] and may be related to F-actin gelation observed in early embryonic extracts [55]. DAPI labeling in (G,I) shows center of blastodisc; astral microtubules in G are not clearly visible as this time point coincides with the cyclical disassembly of this structure after reaching the cortex [16], [56]. (J,K) In situ hybridizations to detect germ plasm RNAs nanos (J) and dead end (K) in motley mutant embryos, showing defects in germ plasm segregation similar to those observed when visualizing GP RNPs with anti-NMII-p antibodies (compare to wild-type in Figure S5D, S5E; GP RNP recruitment in wild-type furrows can be detected as early as 30 mpf [16]). |