Fig. 5
|
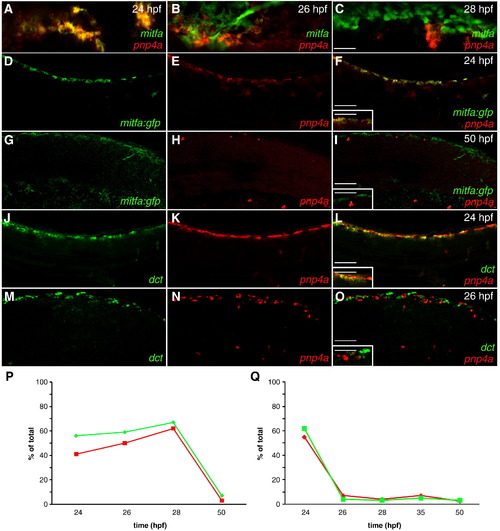
Iridoblast marker co-localizes with melanoblast markers. (A–O) Confocal images collected from lateral aspect of anterior tail region of fixed zebrafish, 20x. (A–C) Cells co-stained with mitfa riboprobe (red) and pnp4a riboprobe (green) reveal a considerable overlap at 24 hpf (A) and a diminishing overlap as development proceeds (B,C). mitfa:gfp transgenic reveals that mitfa+ cells overlap with pnp4a expression at 24 hpf (D–F) and resolve at 50 hpf (G–I). (D,G) mitfa:gfp (E,H) pnp4a. (F,I) Color merged: green: GFP expression, red: pnp4a mRNA (inset 40x). Wild-type embryos reveal that dct+ cells overlap with pnp4a expression at 24 hpf (J–L) then resolve at 26 hpf (M–O). (J,M) dct (K,N) pnp4a. (L,O) Color merged: green: dct mRNA, red: pnp4a mRNA (inset 40x). (P,Q) Percent of overlap between chromatoblast markers (see Table 1). (P) Green line = % of mitfa:gfp+ cells that are mitfa:gfp+/pnp4a+. Red line = % of pnp4a+ cells that are mitfa:gfp+/pnp4a+. (Q) Green line = % of dct+ cells that are dct+/pnp4a+. Red line - % of pnp4a+ cells that are dct+/pnp4a+. Scale bars: (A–C) 40 μm; (D–O) 60 μm; (F,I,L,O inset) 30 μm. |
| Genes: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage Range: | Prim-5 to Long-pec |
Reprinted from Developmental Biology, 344(1), Curran, K., Lister, J.A., Kunkel, G.R., Prendergast, A., Parichy, D.M., and Raible, D.W., Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest, 107-118, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.