- Title
-
The transcription factor Jun is necessary for optic nerve regeneration in larval zebrafish
- Authors
- Sarich, S.C., Sreevidya, V.S., Udvadia, A.J., Svoboda, K.R., Gutzman, J.H.
- Source
- Full text @ PLoS One
|
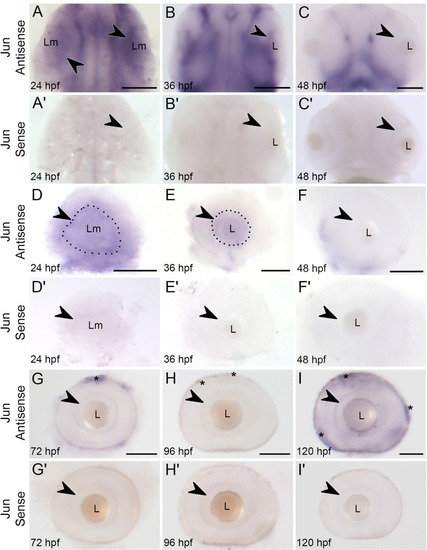
Whole mount |
|
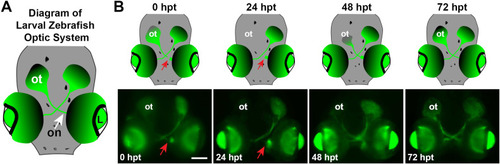
Larval zebrafish optic system, optic nerve transection, and axon regeneration timeline. |
|
( |
|
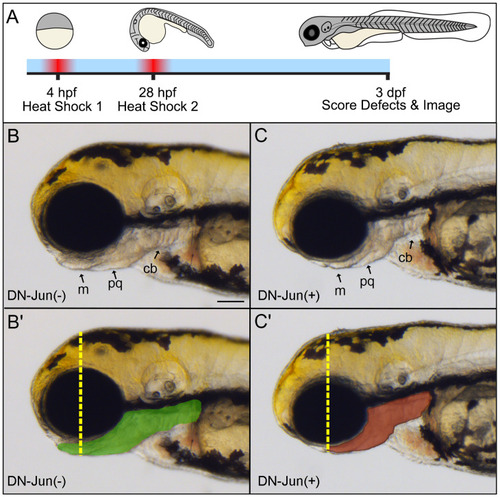
Early induction of DN-Jun results in developmental defects associated with |
|
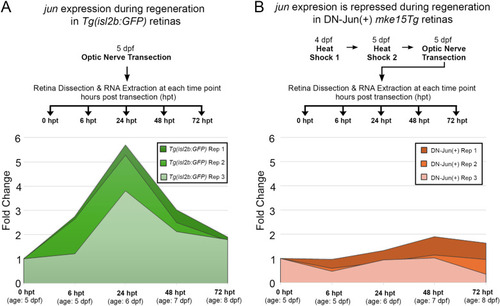
Induction of DN-Jun inhibits endogenous |
|
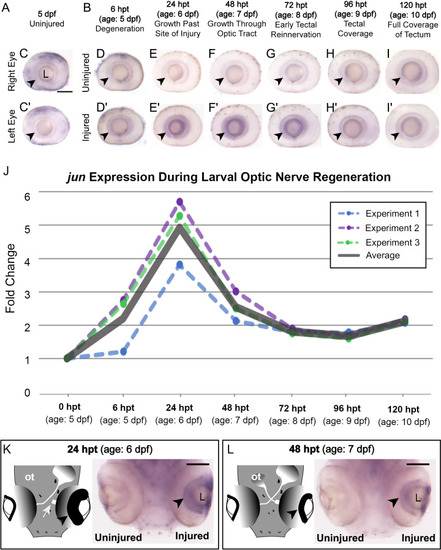
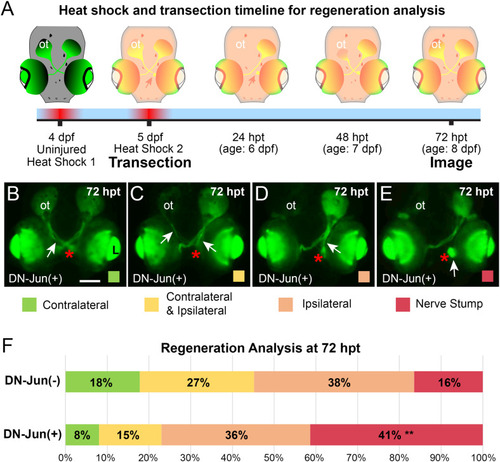
Induction of DN-Jun diminishes capacity for optic nerve regeneration. |
|
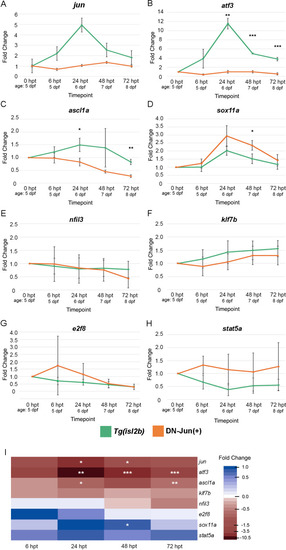
Putative Jun targets display varied expression patterns during larval optic nerve regeneration in control and Jun knockdown conditions. ( |