- Title
-
Common and divergent gene regulatory networks control injury-induced and developmental neurogenesis in zebrafish retina
- Authors
- Lyu, P., Iribarne, M., Serjanov, D., Zhai, Y., Hoang, T., Campbell, L.J., Boyd, P., Palazzo, I., Nagashima, M., Silva, N.J., Hitchcock, P.F., Qian, J., Hyde, D.R., Blackshaw, S.
- Source
- Full text @ Nat. Commun.
|
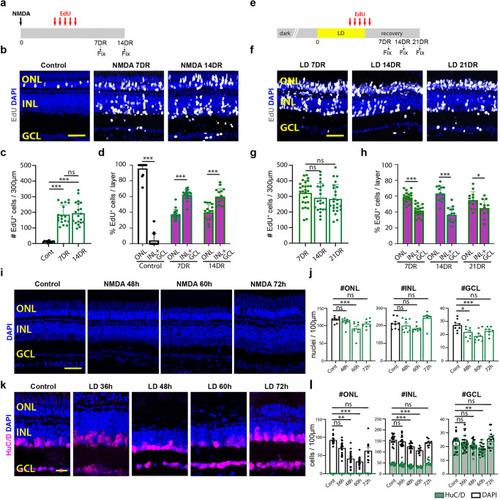
Comparison of NMDA-induced and light-induced retinal damage. |
|
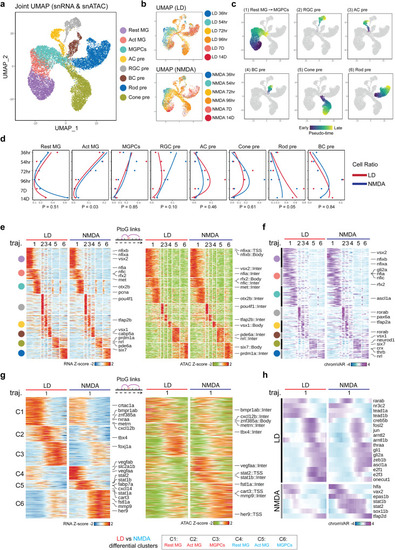
Shared and differential patterns of gene expression and chromatin accessibility data observed in MG-derived cells following LD and NMDA treatment. |
|
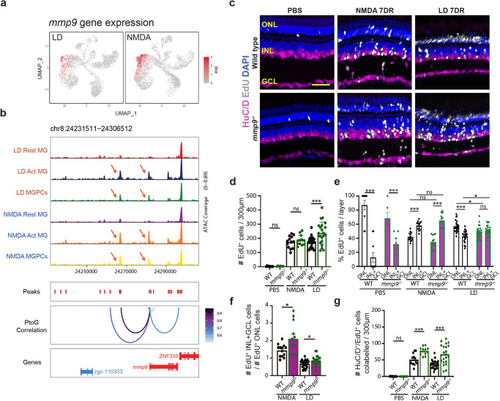
Mmp9 selectively inhibits generation of inner retinal neurons from MGPCs. |
|
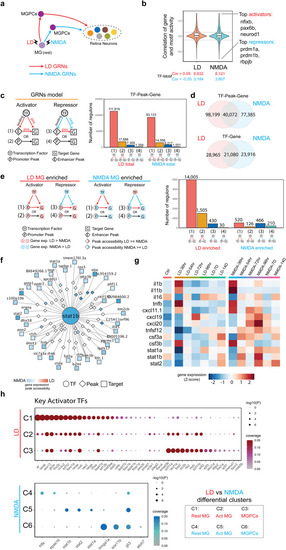
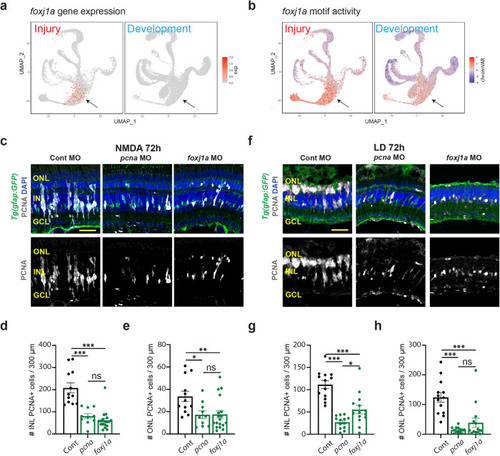
Transcription factors controlling differential gene expression in MG and MGPC following LD and NMDA treatment. |
|
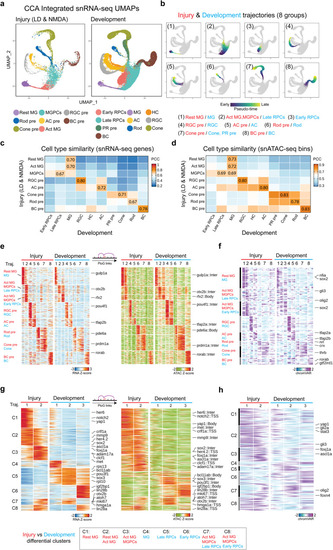
Shared and differential features of MG-derived cells between injury and development datasets |
|
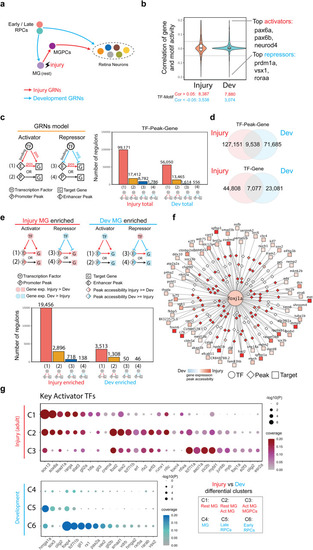
Transcription factors controlling differential expression genes in MGPC in injured retina and progenitor cells in developing retina. |
|
|
|
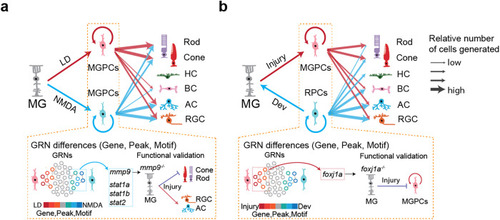
Schematic summary of key findings from this study. |