- Title
-
Adhesion GPCR Gpr126 (Adgrg6) Expression Profiling in Zebrafish, Mouse, and Human Kidney
- Authors
- Cazorla-Vázquez, S., Kösters, P., Bertz, S., Pfister, F., Daniel, C., Dedden, M., Zundler, S., Jobst-Schwan, T., Amann, K., Engel, F.B.
- Source
- Full text @ Cells
|
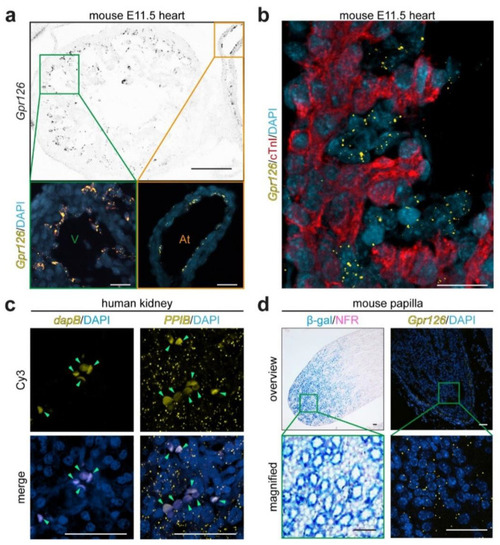
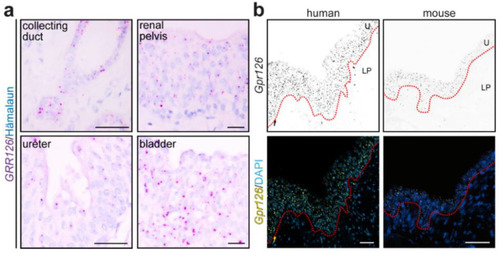
Validation of RNAscope® for mouse and human kidneys. ( |
|
|
|
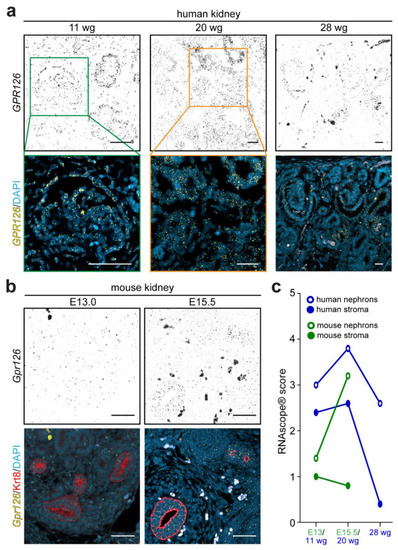
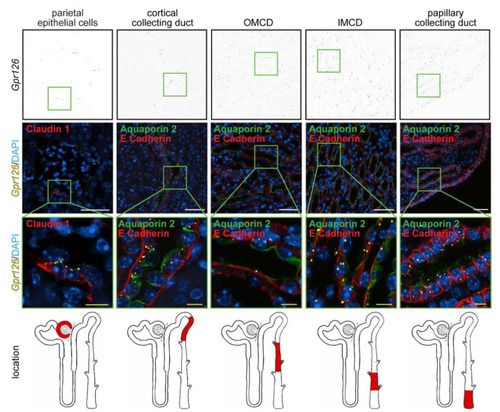
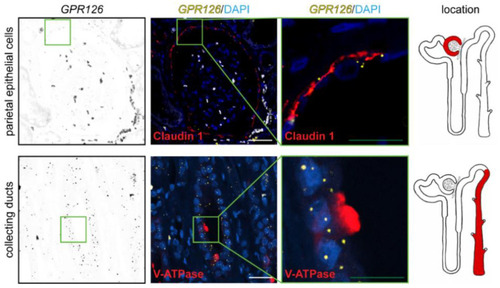
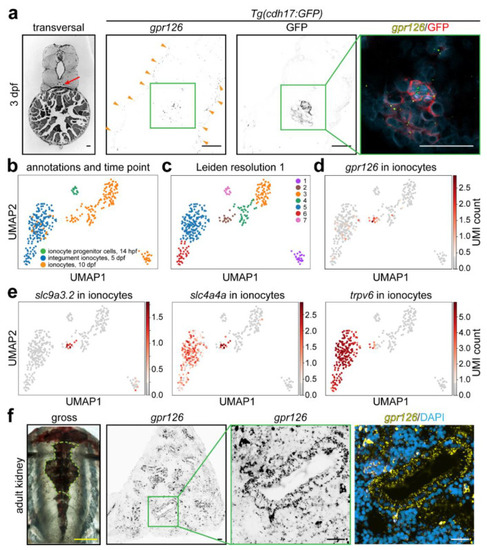
Renal expression of |
|
|
|
|
|
|