- Title
-
Using Zebrafish to Dissect the Interaction of Mycobacteria with the Autophagic Machinery in Macrophages
- Authors
- Muñoz-Sánchez, S., Varela, M., van der Vaart, M., Meijer, A.H.
- Source
- Full text @ Biology (Basel)
|
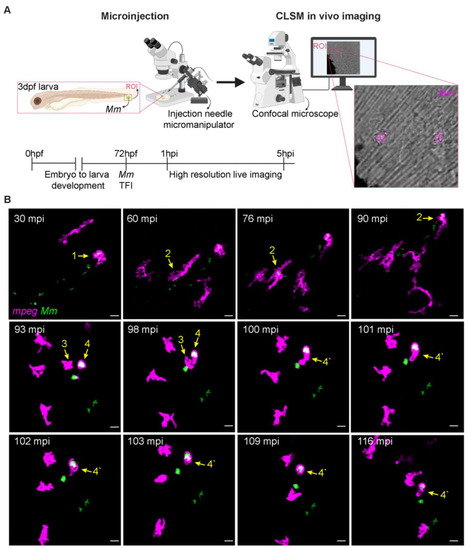
TFI high-resolution live imaging. ( |
|
|
|
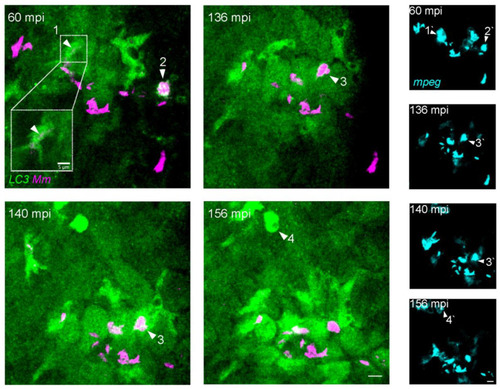
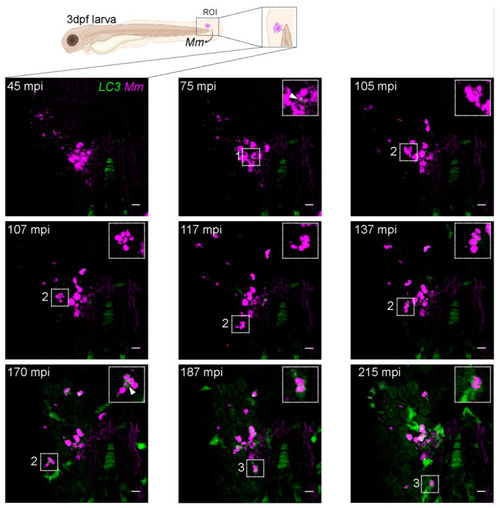
Intracellular dynamics of LC3 association with |
|
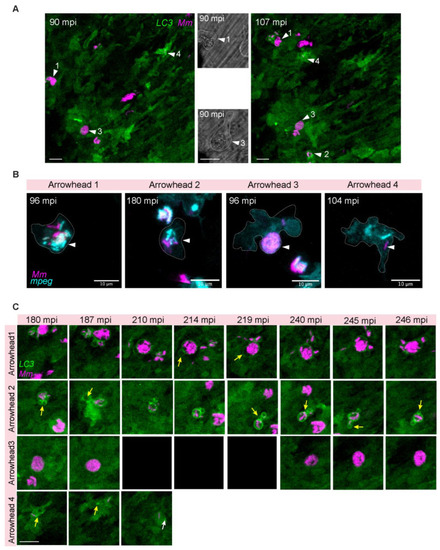
Heterogenous dynamic morphologies of LC3-positive |
|
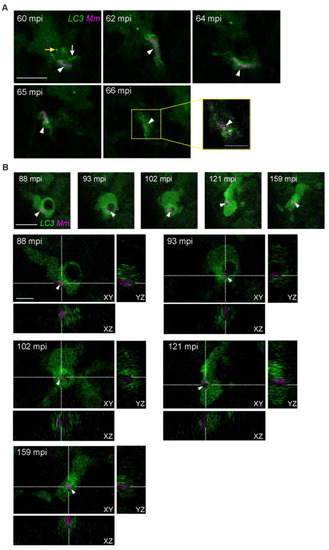
Early dissemination of |
|
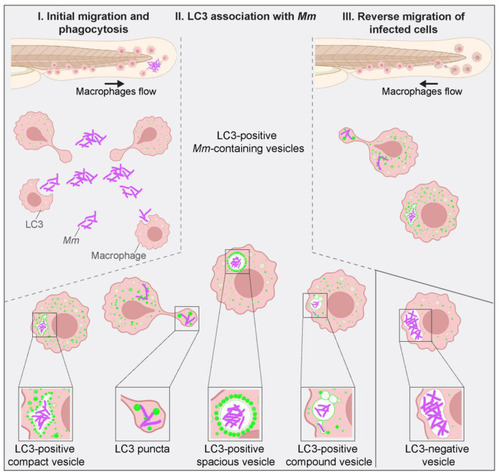
Schematic overview of LC3-associations with |