- Title
-
Svep1 is a binding ligand of Tie1 and affects specific aspects of facial lymphatic development in a Vegfc-independent manner
- Authors
- Hußmann, M., Schulte, D., Weischer, S., Carlantoni, C., Nakajima, H., Mochizuki, N., Stainier, D.Y.R., Zobel, T., Koch, M., Schulte-Merker, S.
- Source
- Full text @ Elife
|
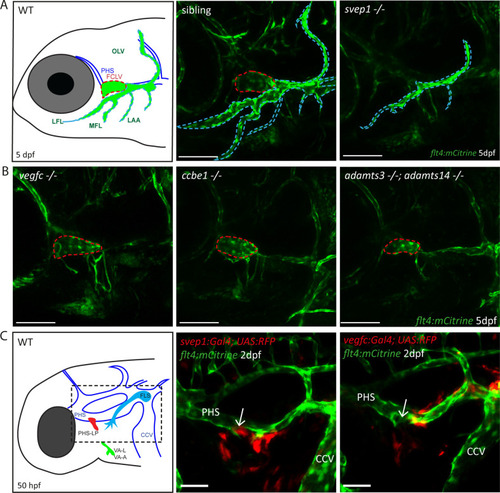
(A) Schematic representation of facial lymphatic network at 5 dpf and maximum intensity projection of confocal images of flt4:mCitrine positive svep1 mutants (n = 10) and siblings (n = 6), highlighting facial lymphatic structures at 5 dpf. Scale bar = 100 µm. Note the absence of the FCLV (red dotted line) in svep1 mutants whereas other facial lymphatic structures are less strongly affected (OLV, LFL, MFL, and LAA marked by blue dotted lines). (B) Confocal images of flt4:mCitrine positive facial lymphatics in vegfc (n = 19), ccbe1 (n = 5), and adamts3;adamts14 (n = 2) mutants at 5 dpf. Scale bar = 100 µm. (C) Confocal images of svep1 and vegfc expression domains during sprouting from the PHS at 2 dpf, with schematic representation of different lymphatic progenitor populations. svep1 is expressed in close proximity to sprouting PHS-LPs, while vegfc expressing cells are more concentrated on the LECs arising from the CCV. Arrows point to sprouting PHS-LP. Scale bar = 50 µm. Expression patterns were confirmed in six embryos each (Figure 1—figure supplement 3). CCV, common cardinal vein; dpf, days post-fertilization; FCLV, facial collecting lymphatic vessel; FLS, facial lymphatic sprout; hpf, hours post-fertilization; LAA, lymphatic branchial arches; LEC, lymphatic endothelial cell; LFL, lateral facial lymphatic; MFL, medial facial lymphatic; OLV, otolithic lymphatic vessel; PHS, primary head sinus; PHS-LP, primary head sinus lymphatic progenitor; VA, ventral aorta; VA-A, ventral aorta angioblast; VA-L, ventral aorta lymphangioblast; WT, wildtype.
|
|
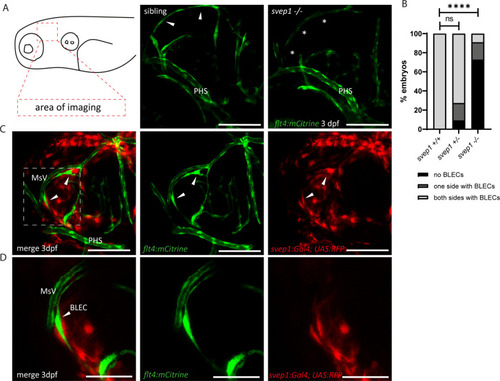
(A) Confocal images of sprouting BLECs, marked by flt4:mCitrine, at 3 dpf in svep1 mutants and siblings. Asterisks mark missing BLECs in svep1 mutants. Scale bar = 100 µm. (B) Quantification of BLECs at 3 dpf on each side of the embryo showed that svep1 mutants have significantly less BLECs on one or both sides of the brain hemispheres compared to siblings. For statistical analysis, no BLECs were counted as 0, BLECs being present on only one hemisphere as 1, whereas BLECs being detectable on both brain hemispheres were included as 2, for each embryo (svep1+/+: n = 10; svep1+/−: n = 12; svep1−/−: n = 12). Mann–Whitney test was applied for statistical analysis. Values are presented as means ± standard deviation (SD), ****p < 0.0001, ns = not significant. Scale bar = 100 µm. (C) Confocal images of svep1:Gal4; UAS:RFP, showing svep1 expression immediately adjacent to BLECs, marked by arrowheads, at 3 dpf. Scale bar = 100 µm. (D) Magnification and reduced stack numbers of boxed area in (C). Arrowhead marks BLEC. Scale bar = 50 µm. BLEC, brain lymphatic endothelial cell; dpf, days post-fertilization; MsV, mesencephalic vein; PHS, primary head sinus;.
|
|
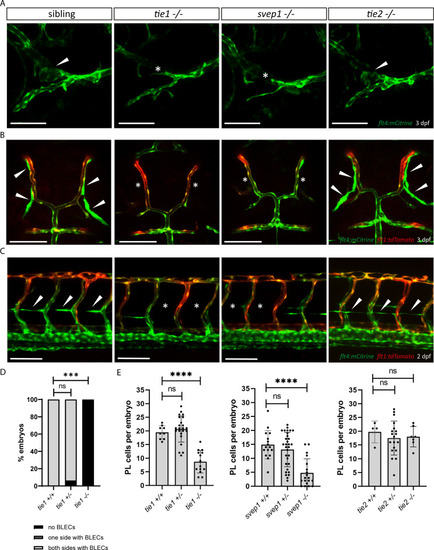
(A) Facial lymphatics at 3 dpf in flt4:mCitrine positive tie1, svep1 and tie2 mutants and sibling embryos (lateral view). Arrowheads point to FCLV and asterisks indicate the absence of FCLV. Scale bar = 100 µm. (B) flt4:mCitrine; flt1:tdTomato positive dorsal head vasculature in tie1, svep1, and tie2 mutants and in siblings at 3 dpf (dorsal view). In svep1 and tie1 mutants (but not in tie2 mutants) the presence of BLECs is strongly reduced. Arrowheads point to BLECs and asterisks indicate areas lacking BLECs. Scale bar = 100 µm. (C) Confocal images of PL cells, indicated by arrowheads, at 2 dpf in flt4:mCitrine; flt1:tdTomato positive tie1, svep1, and tie2 mutants and siblings, showing reduced PL numbers in svep1 and tie1 mutants. Asterisks indicate missing PLs. Scale bar = 100 µm. (D) Quantification of the presence of BLECs in tie1 mutants compared to siblings. (tie1+/+: n = 6; tie1+/−: n = 16; tie1−/−: n = 10) Mann–Whitney test was applied for statistical analysis. ***p = 0.001, ns = not significant. (E) Quantification of PL cell numbers in tie1 (tie1+/+: n = 9; tie1+/−: n = 23; tie1−/−: n = 14), svep1 (svep1+/+: n = 16; svep1+/−: n = 31; svep1−/−: n = 19), and tie2 (tie2+/+: n = 17; tie2+/−: n = 27; tie2−/−: n = 16) mutants compared to siblings. Mann–Whitney test was applied for statistical analysis. Values are presented as means ± standard deviation (SD), ****p < 0.0001, ns = not significant; BLEC, brain lymphatic endothelial cell; dpf, days post-fertilization; FCLV, facial collecting lymphatic vessel; PL, parachordal lymphangioblast.
|
|
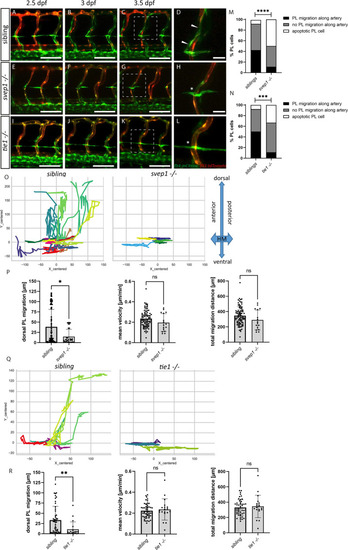
(A–L) Still frames from confocal time-lapse imaging of embryos in a flt4:mCitrine; flt1:tdTomato transgenic background. (A–D) PL migration (indicated by arrowheads) of sibling embryo along aISV from 2.5 to 3.5 dpf. (E–H) Failed PL migration (indicated by asterisk) of svep1 mutants and (I–L) tie1 mutants along artery from 2.5 to 3.5 dpf. (M, N) Classification of PL migration along arteries. Statistical analysis was performed using Mann–Whitney test comparing the % of PL migration along arteries in each sibling and mutant embryo (sibling: n = 96 PLs in 18 embryos; svep1−/−: n = 36 PLs in 15 embryos; siblings: n = 52 PLs in 14 embryos; tie1−/−: n = 28 PLs in 10 embryos); ****p < 0.0001, ***p = 0.0003. (O, Q) Representative cell tracking routes (tracks centred to origin) of single PL cells marked by different colours in siblings (n = 17 PLs in 4 embryos; n = 7 in 2 embryos), tie1−/− (n = 5 PLs in 2 embryos) and svep1−/− (n = 6 PLs in 3 embryos). (P, R) Quantification of dorsal and ventral PL migration (delta Y migration distance), mean velocity and total migration distance in svep1 and tie1 mutants compared to sibling embryos excluding apoptotic PLs quantified in (M, N) revealed decreased migration in dorsal and ventral direction in svep1 (*p = 0.0148) as well as tie1 mutants (**p = 0.0023). ns = not significant; aISV, arterial intersegmental vessel; dpf, days post fertilization; HM, horizontal myoseptum; PL, parachordal lymphangioblast. Scale bar = 100 µm (D, H, L = 25 µm).
|
|
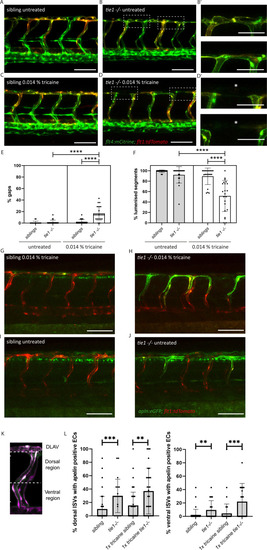
(A, B) Confocal images of sibling and tie1 mutant embryos at 2 dpf in a flt4:mCitrine and flt1:tdTomato transgenic background. (B’) Magnification and reduced stack of boxed area in (B). (C, D) Confocal images of sibling and tie1 mutant embryos treated with 0.014% tricaine from 30 until 48 hpf. Asterisks indicate incompletely formed DLAV segments. (D’) Magnification and reduced stack numbers of boxed area in (D). (E) Quantification of gaps in the DLAV in sibling and tie1 mutants that were either untreated or treated with 0.014% tricaine revealed significant increase of gaps in the DLAV in tie1 mutants. (F) Quantification of lumenized trunk segments of the DLAV in siblings and tie1 mutants, either untreated or treated with 0.014% tricaine (siblings untreated: n = 16; tie1−/− untreated: n = 20; siblings treated with 0.014% tricaine: n = 20; tie1−/− treated with 0.014% tricaine: n = 22), revealed significant decrease of lumenized segment numbers in the DLAV in tie1 mutants. Mann–Whitney test was applied for statistical analysis. (G, H) apelin:eGFP and flt1:tdTomato expression in 48-hpf-old embryos after tricaine treatment from 30 to 48 hpf and (I, J) in untreated conditions. (K) Maximum intensity projection of an aISV at 48 hpf, highlighting the ventral and dorsal region used for further quantifications in (J) adapted from Figure 5J of Coxam et al., 2022. (L) Quantification of ISVs with apelin expression in dorsal and ventral parts of the ISVs. Dorsal part was counted from DLAV until midline region. Lateral region was counted from midline region onwards in ventral direction. tie1 mutants showed significant increase of apelin positive ECs compared to siblings in untreated (dorsal: ***p = 0.0001; ventral: **p = 0.0028) and treated with 0.014% tricaine conditions (dorsal: **p = 0.0033; ventral: ***p = 0.0002) (siblings untreated: n = 53; tie1−/− untreated: n = 21; siblings treated with 0.014% tricaine: n = 66; tie1−/− treated with 0.014% tricaine: n = 28). Mann–Whitney test was applied for statistical analysis. Values are presented as means ± standard deviation (SD). ****p < 0.0001. Scale bar = 100 µm. hpf, hours post-fertilization; ISV, intersegmental vessel; DLAV, dorsal longitudinal anastomotic vessel; dpf, days post-fertilization.
|
|
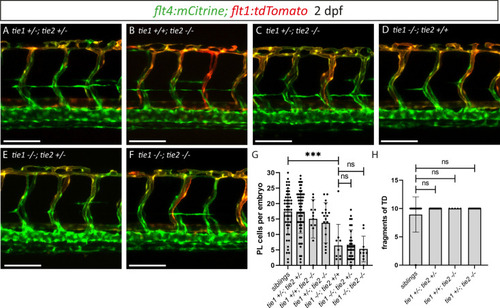
(A–F) Confocal images of blood and lymphatic vasculature in the trunk of 2 dpf old embryos derived from tie1; tie2 double heterozygous parents, showing no genetic interaction between tie1 and tie2. (G) Quantification of PLs at 2 dpf and of thoracic duct fragments at 5 dpf (siblings: n = 50; tie1+/−; tie2+/−: n = 62; tie1+/+; tie2−/−: n = 13; tie1+/−; tie2−/−: n = 20; tie1−/−; tie2+/+: n = 10; tie1−/−; tie2+/−: n = 32; tie1−/−; tie2−/−: n = 10). (H) TD fragments were counted over the anterior-most 10 somites (siblings: n = 47; tie1+/−; tie2+/−: n = 34; tie1+/+; tie2−/−: n = 5; tie1+/−; tie2−/−: n = 16). Mann–Whitney test was applied for statistical analysis. ***p = 0.0002, ns = not significant. Scale bar = 100 µm. dpf, days post-fertilization; PL, parachordal lymphangioblast; TD, thoracic duct.
|
|
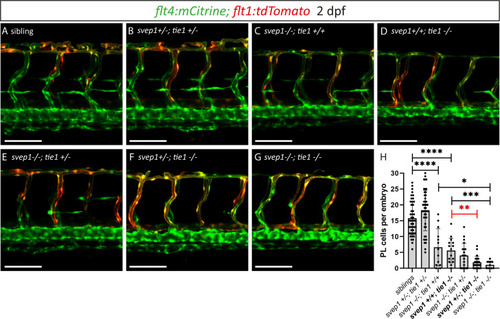
(A–G) Confocal images of blood and lymphatic vasculature in the trunk of 2-dpf-old embryos derived from svep1; tie1 double heterozygous fish, showing severely reduced PL numbers in svep1; tie1 double mutants and significant decrease of PL cell numbers in svep1+/−; tie1−/− compared to svep1+/+; tie1−/− (**p = 0.0012). (H) Quantification of PL cell numbers at 2 dpf using Mann–Whitney test (siblings: n = 45; svep1+/−; tie1+/−: n = 45; svep1−/−; tie1+/+: n = 13; svep1+/+; tie1−/−: n = 15; svep1−/−; tie1+/−: n = 20; svep1+/−; tie1−/−: n = 21; svep1−/−; tie1−/−: n = 11). Scale bar = 100 µm. Values are presented as means ± standard deviation (SD), ****p < 0.0001, ***p = 0.007, *p = 0.0163, ns = not significant. dpf, days post-fertilization; PL, parachordal lymphangioblast.
|
|
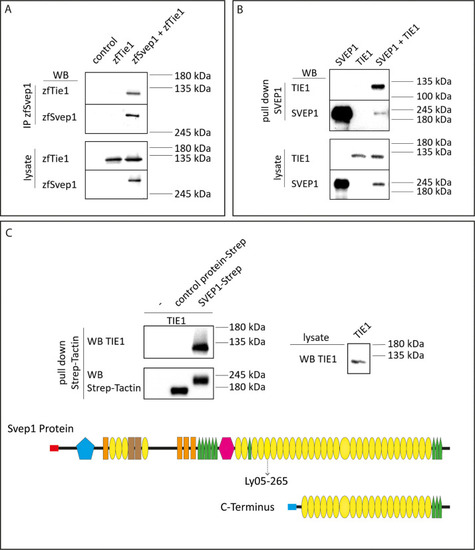
(A) 293T HEK cells were transfected with zebrafish Svep1-HIS (zfSvep1) and zebrafish Tie1-HA (zfTie1). zfSvep1 was immunoprecipitated and associated Tie1 was detected by western blot. (B) Co-immunoprecipitation of C-terminal human SVEP1 co-transfected in 293T HEK cells with human TIE1. (C) Pull-down of recombinant C-terminal human SVEP1-Strep-tag II protein, which was incubated with TIE1 transfected 293T HEK cell lysates, shows binding of TIE1. Protein structure with all domains indicated and C-terminal part used for pull-down assays (adapted from Figure 2F of Karpanen et al., 2017, published under the CC BY-NC 4.0 license, https://creativecommons.org/licenses/by-nc/4.0/). It is not covered by the CC-BY 4.0 license and further reproduction of this panel would need to follow the terms of the CC BY-NC 4.0 license. Ly05-265 indicates position of stop codon in the zebrafish hu6985 allele (Karpanen et al., 2017), suggesting that the protein domains C-terminal to the nonsense allele are critical for function. Red and blue rectangle: signal peptide; blue pentagon: von Willebrand factor type A domain (vWF); orange rectangle: ephrin-receptor like domain; brown rectangle: Hyalin repeat; yellow ovals: SUSHI repeat; green pentagons: epidermal growth factor (EGF)-like and calcium-binding EGF-like domains; and pink hexagon: pentraxin domain (PTX). © 2017, Karpanen et al. (C) Pull -down of recombinant C-terminal human SVEP1-Strep-tag II protein, which was incubated with TIE1 transfected 293 T HEK cell lysates, shows binding of TIE1. Protein structure with all domains indicated and C-terminal part used for pull down assays (adapted from Figure 2F of Karpanen et al., 2017, published under the CC BY-NC 4.0 license (https://creativecommons.org/licenses/by-nc/4.0/). It is not covered by the CC-BY 4.0 license and further reproduction of this panel would need to follow the terms of the CC BY-NC 4.0 license)
|