- Title
-
ERBB and P-glycoprotein inhibitors break resistance in relapsed neuroblastoma models through P-glycoprotein
- Authors
- Rösch, L., Herter, S., Najafi, S., Ridinger, J., Peterziel, H., Cinatl, J., Jones, D.T.W., Michaelis, M., Witt, O., Oehme, I.
- Source
- Full text @ Mol. Oncol.
|
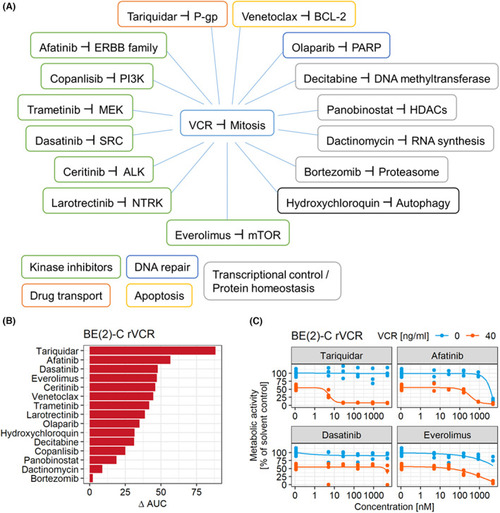
Functional screen identifies ERBB4 and P‐gp inhibitors to break resistance. (A) Schematic representation of the drugs and their targets. Drug classes are indicated. (B) A metabolic activity screen read‐out (CellTiterGlo) of combination treatment of VCR with 15 anticancer drugs (14 clinically approved) was performed with BE(2)‐C rVCR. The cells were treated with 0, 5, 28, 158, 889, and 5000 n |
|
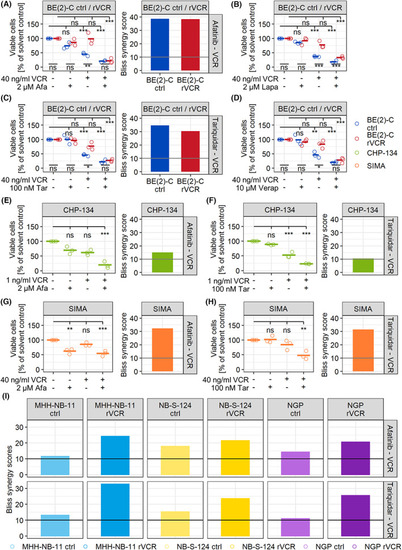
Inhibitors of the ERBB family and P‐gp break resistance in BE(2)‐C rVCR. (A–D) BE(2)‐C control and rVCR cells were treated with 40 ng·mL−1 VCR and the indicated concentrations of the ERBB inhibitors afatinib (A) and lapatinib (B) or the P‐gp inhibitors tariquidar (C) or verapamil (D) for 48 h. (E, F) CHP‐134 cells were treated with 1 ng·mL−1 VCR and 2 μ |
|
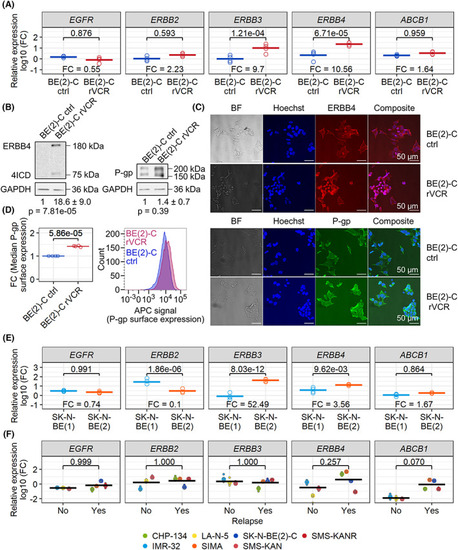
ERBB3, ERBB4, and P‐gp/ |
|
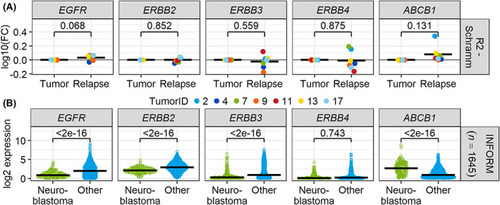
ABCB1 is upregulated in neuroblastoma relapsed tumors. (A) Gene expression data of 18 tumor samples from the dataset by Schramm et al. ( |
|
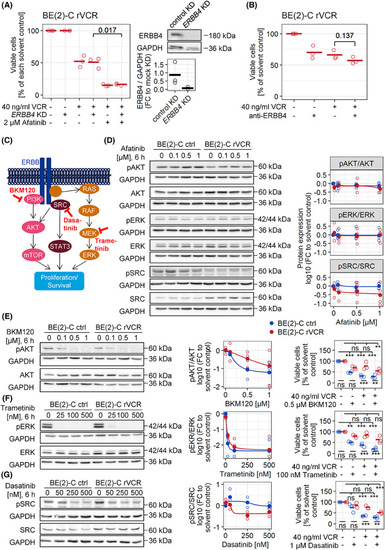
Afatinib breaks VCR resistance independent of ERBB4. (A) |
|
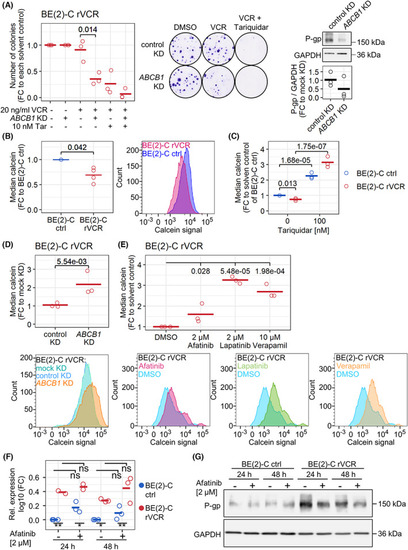
P‐gp mediates resistance in BE(2)‐C rVCR. (A) a colony assay was performed after |
|
Combination of VCR with afatinib or tariquidar induces apoptosis and reduces tumor volume |